1BCI
 
 | | C2 DOMAIN OF CYTOSOLIC PHOSPHOLIPASE A2, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, CYTOSOLIC PHOSPHOLIPASE A2 | | Authors: | Xu, G.Y, Mcdonagh, T, Yu, H.A, Nalefski, E.A, Clark, J.D, Cumming, D.A. | | Deposit date: | 1998-04-30 | | Release date: | 1998-11-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and membrane interactions of the C2 domain of cytosolic phospholipase A2.
J.Mol.Biol., 280, 1998
|
|
1EXG
 
 | | SOLUTION STRUCTURE OF A CELLULOSE BINDING DOMAIN FROM CELLULOMONAS FIMI BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | EXO-1,4-BETA-D-GLYCANASE | | Authors: | Xu, G.-Y, Ong, E, Gilkes, N.R, Kilburn, D.G, Muhandiram, D.R, Harris-Brandts, M, Carver, J.P, Kay, L.E, Harvey, T.S. | | Deposit date: | 1995-03-14 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1EXH
 
 | | SOLUTION STRUCTURE OF A CELLULOSE BINDING DOMAIN FROM CELLULOMONAS FIMI BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | EXO-1,4-BETA-D-GLYCANASE | | Authors: | Xu, G.-Y, Ong, E, Gilkes, N.R, Kilburn, D.G, Muhandiram, D.R, Harris-Brandts, M, Carver, J.P, Kay, L.E, Harvey, T.S. | | Deposit date: | 1995-03-14 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
2VVZ
 
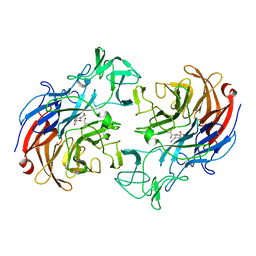 | | Structure of the catalytic domain of Streptococcus pneumoniae sialidase NanA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CHLORIDE ION, SIALIDASE A | | Authors: | Xu, G, Li, X, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Catalytic Domain of Streptococcus Pneumoniae Sialidase Nana.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2W20
 
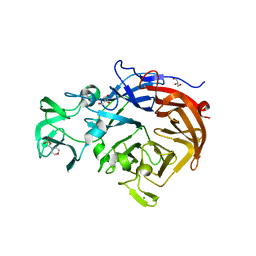 | | Structure of the catalytic domain of the native NanA sialidase from Streptococcus pneumoniae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Xu, G, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-10-21 | | Release date: | 2008-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of the Catalytic Domain of Streptococcus Pneumoniae Sialidase Nana.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2W38
 
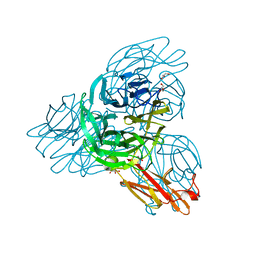 | | Crystal structure of the pseudaminidase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, SIALIDASE | | Authors: | Xu, G, Ryan, C, Kiefel, M.J, Wilson, J.C, Taylor, G.L. | | Deposit date: | 2008-11-07 | | Release date: | 2008-12-23 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on the Pseudomonas Aeruginosa Sialidase-Like Enzyme Pa2794 Suggest Substrate and Mechanistic Variations.
J.Mol.Biol., 386, 2009
|
|
2VW1
 
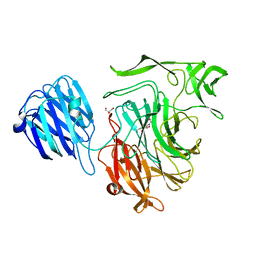 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
2VW2
 
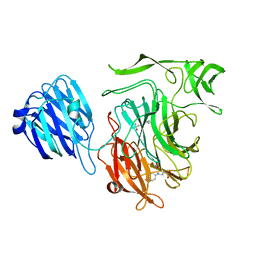 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
2VW0
 
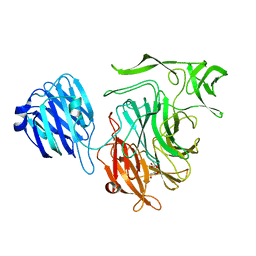 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
1IL6
 
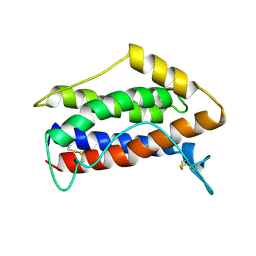 | | HUMAN INTERLEUKIN-6, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INTERLEUKIN-6 | | Authors: | Xu, G.Y, Yu, H.A, Hong, J, Stahl, M, Mcdonagh, T, Kay, L.E, Cumming, D.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of recombinant human interleukin-6.
J.Mol.Biol., 268, 1997
|
|
1HY8
 
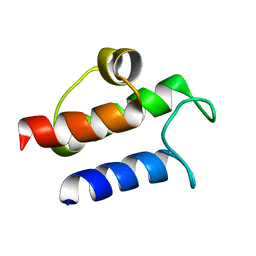 | | SOLUTION STRUCTURE OF B. SUBTILIS ACYL CARRIER PROTEIN | | Descriptor: | ACYL CARRIER PROTEIN | | Authors: | Xu, G.-Y, Tam, A, Lin, L, Hixon, J, Fritz, C.C, Power, R. | | Deposit date: | 2001-01-18 | | Release date: | 2002-01-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of B. subtilis acyl carrier protein.
Structure, 9, 2001
|
|
1I4E
 
 | | CRYSTAL STRUCTURE OF THE CASPASE-8/P35 COMPLEX | | Descriptor: | Caspase-8, Early 35 kDa protein | | Authors: | Xu, G, Cirilli, M, Huang, Y, Rich, R.L, Myszka, D.G, Wu, H. | | Deposit date: | 2001-02-20 | | Release date: | 2001-03-28 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Covalent inhibition revealed by the crystal structure of the caspase-8/p35 complex.
Nature, 410, 2001
|
|
8YZN
 
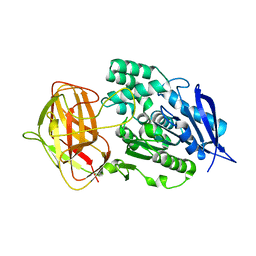 | |
8YZO
 
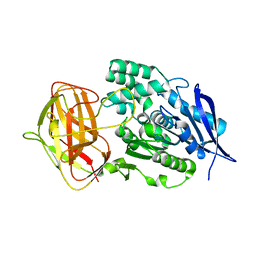 | |
5GGM
 
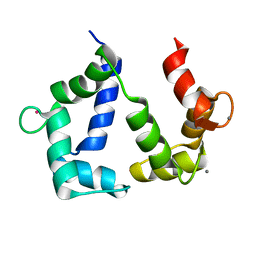 | | The NMR structure of calmodulin in CTAB reverse micelles | | Descriptor: | CALCIUM ION, Calmodulin, TERBIUM(III) ION | | Authors: | Xu, G, Cheng, K, Wu, Q, Liu, M, Li, C. | | Deposit date: | 2016-06-16 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of calmodulin in CTAB reverse micelles
To Be Published
|
|
7W3U
 
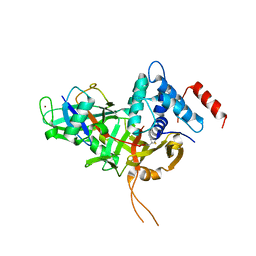 | | USP34 catalytic domain in complex with UbPA | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION, ... | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
8IF5
 
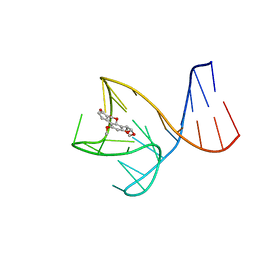 | | AFB1-AF26 APTAMER COMPLEX | | Descriptor: | AFB1 DNA aptamer (26-MER), AFLATOXIN B1 | | Authors: | Xu, G.H, Wang, C, Li, C.G. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer.
Nucleic Acids Res., 51, 2023
|
|
5JFI
 
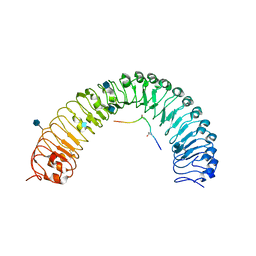 | | Crystal structure of a TDIF-TDR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CLE41, Leucine-rich repeat receptor-like protein kinase TDR | | Authors: | Xu, G, Li, Z. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Crystal structure of a TDIF-TDR complex
To Be Published
|
|
1YMW
 
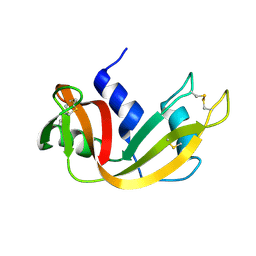 | | The study of reductive unfolding pathways of RNase A (Y92G mutant) | | Descriptor: | Ribonuclease pancreatic | | Authors: | Xu, G, Narayan, M, Kurinov, I, Ripoll, D.R, Welker, E, Khalili, M, Ealick, S.E, Scheraga, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2006-01-31 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A localized specific interaction alters the unfolding pathways of structural homologues.
J.Am.Chem.Soc., 128, 2006
|
|
1YMN
 
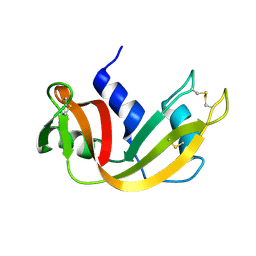 | | The study of reductive unfolding pathways of RNase A (Y92L mutant) | | Descriptor: | Ribonuclease pancreatic | | Authors: | Xu, G, Narayan, M, Kurinov, I, Ripoll, D.R, Welker, E, Khalili, M, Ealick, S.E, Scheraga, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A localized specific interaction alters the unfolding pathways of structural homologues.
J.Am.Chem.Soc., 128, 2006
|
|
1YMR
 
 | | The study of reductive unfolding pathways of RNase A (Y92A mutant) | | Descriptor: | Ribonuclease pancreatic | | Authors: | Xu, G, Narayan, M, Kurinov, I, Ripoll, D.R, Welker, E, Khalili, M, Ealick, S.E, Scheraga, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2006-01-31 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A localized specific interaction alters the unfolding pathways of structural homologues.
J.Am.Chem.Soc., 128, 2006
|
|
3RZF
 
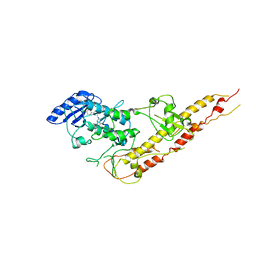 | | Crystal Structure of Inhibitor of kappaB kinase beta (I4122) | | Descriptor: | (4-{[4-(4-chlorophenyl)pyrimidin-2-yl]amino}phenyl)[4-(2-hydroxyethyl)piperazin-1-yl]methanone, MGC80376 protein | | Authors: | Xu, G, Lo, Y.C, Li, Q, Napolitano, G, Wu, X, Jiang, X, Dreano, M, Karin, M, Wu, H. | | Deposit date: | 2011-05-11 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of inhibitor of KappaB kinase Beta.
Nature, 472, 2011
|
|
2IL6
 
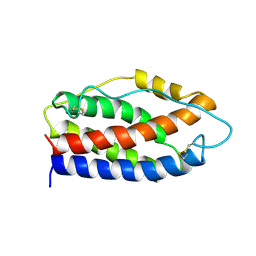 | | HUMAN INTERLEUKIN-6, NMR, 32 STRUCTURES | | Descriptor: | INTERLEUKIN-6 | | Authors: | Xu, G.Y, Yu, H.A, Hong, J, Stahl, M, Mcdonagh, T, Kay, L.E, Cumming, D.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of recombinant human interleukin-6.
J.Mol.Biol., 268, 1997
|
|
3QA8
 
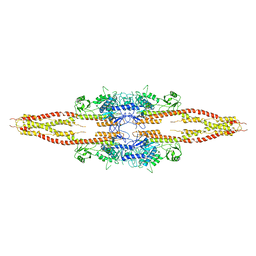 | | Crystal Structure of inhibitor of kappa B kinase beta | | Descriptor: | MGC80376 protein | | Authors: | Xu, G, Lo, Y.C, Li, Q, Napolitano, G, Wu, X, Jiang, X, Dreano, M, Karin, M, Wu, H. | | Deposit date: | 2011-01-10 | | Release date: | 2011-04-06 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of inhibitor of kappa B kinase beta.
Nature, 472, 2011
|
|
7W3R
 
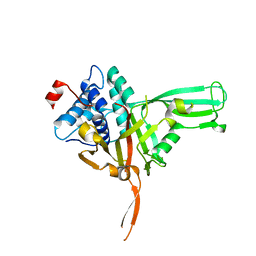 | | USP34 catalytic domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
