2CFR
 
 | |
5WBY
 
 | |
8HQ6
 
 | | KL2 in complex with CRM1-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8HQ3
 
 | | KL1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8UX1
 
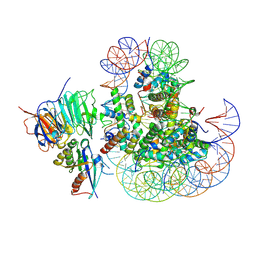 | |
8TYQ
 
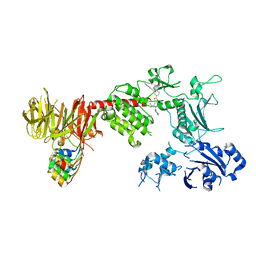 | | Structure of the C-terminal half of LRRK2 bound to GZD-824 (G2019S mutant) | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, Designed Ankyrin Repeats Protein E11, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Villagran-Suarez, A, Sanz-Murillo, M, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8TZE
 
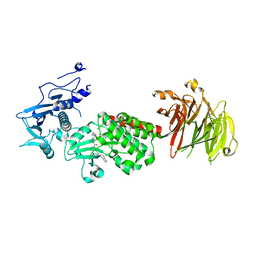 | | Structure of C-terminal half of LRRK2 bound to GZD-824 | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Villagran-Suarez, A, Sanz-Murillo, M, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-26 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8TZH
 
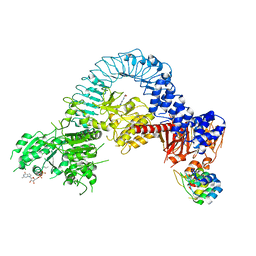 | | Structure of full-length LRRK2 bound to MLi-2 (I2020T mutant) | | Descriptor: | (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}-indazol-3-yl]pyrimidin-4-yl]morpholine, E11 DARPin, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sanz-Murillo, M, Villagran-Suarez, A, Alegrio Louro, J, Leschziner, A. | | Deposit date: | 2023-08-26 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8TZF
 
 | | Structure of full length LRRK2 bound to GZD-824 (I2020T mutant) | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, ... | | Authors: | Villagran-Suarez, A, Sanz-Murillo, M, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-26 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8TZB
 
 | | Structure of the C-terminal half of LRRK2 bound to GZD-824 (I2020T mutant) | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, Leucine-rich repeat serine/threonine-protein kinase 2, designed ankyrin repeat proteins E11 | | Authors: | Villagran-Suarez, A, Sanz-Murillo, M, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-26 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8HQ4
 
 | | B27 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3-[(4-fluorophenyl)carbonylamino]-4-[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]benzoic acid, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Lei, Y, Sun, Q. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | B27 in complex with CRM1-Ran-RanBP1
To Be Published
|
|
8HQ5
 
 | | G6 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3-[(4-bromophenyl)carbonylamino]-4-[4-(5-chloranyl-2-methyl-phenyl)piperazin-1-yl]benzoic acid, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | F1 in complex with CRM1-Ran-RanBP1
To Be Published
|
|
8HUG
 
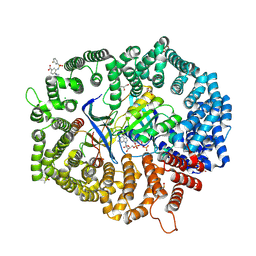 | | F1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 4-[4-(3-chlorophenyl)piperazin-1-yl]-3-[(3-fluorophenyl)sulfonylamino]benzoic acid, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Hidden Pocket beneath the NES Groove by Novel Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8HUF
 
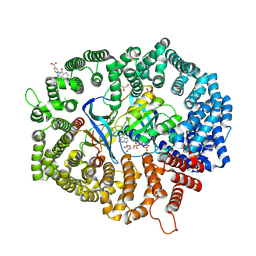 | | B28 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3-[(4-chlorophenyl)carbonylamino]-4-[4-(2,5-dimethylphenyl)piperazin-1-yl]benzoic acid, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of a Hidden Pocket beneath the NES Groove by Novel Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8U4N
 
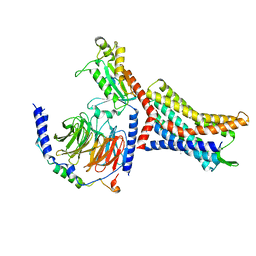 | | Structure of Apo CXCR4/Gi complex | | Descriptor: | C-X-C chemokine receptor type 4, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization
Biorxiv, 2024
|
|
8U4R
 
 | |
8U4T
 
 | |
8U4P
 
 | |
8U4S
 
 | |
8U4Q
 
 | |
8I8Y
 
 | | A mutant of the C-terminal complex of proteins 4.1G and NuMA | | Descriptor: | Engineered protein | | Authors: | Hu, X. | | Deposit date: | 2023-02-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Combined prediction and design reveals the target recognition mechanism of an intrinsically disordered protein interaction domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5WBU
 
 | |
8ILL
 
 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH5.6 | | Descriptor: | CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, green fluorescent protein | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
8ILK
 
 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH8.5 | | Descriptor: | CHLORIDE ION, Green FLUORESCENT PROTEIN | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
8V5Y
 
 | |
