3I89
 
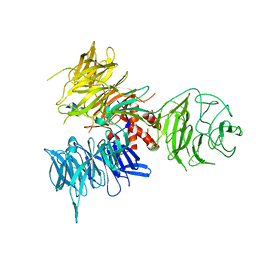 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDR22 | | Descriptor: | DNA damage-binding protein 1, WD repeat-containing protein 22 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-09 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2RMW
 
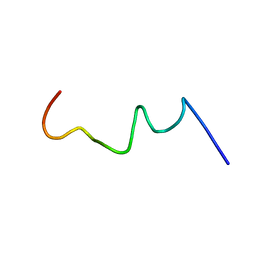 | |
3IK7
 
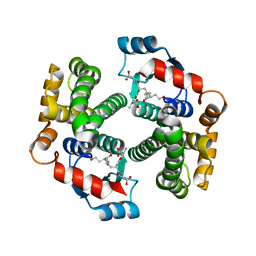 | | Human glutathione transferase a4-4 with GSDHN | | Descriptor: | (S)-2-amino-5-((R)-1-(carboxymethylamino)-3-((3S,4R)-1,4-dihydroxynonan-3-ylthio)-1-oxopropan-2-ylamino)-5-oxopentanoic acid, Glutathione S-transferase A4, SULFATE ION | | Authors: | Balogh, L.M, Le Trong, I, Atkins, W.M, Stenkamp, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Substrate specificity combined with stereopromiscuity in glutathione transferase A4-4-dependent metabolism of 4-hydroxynonenal.
Biochemistry, 49, 2010
|
|
3EVQ
 
 | |
2RNL
 
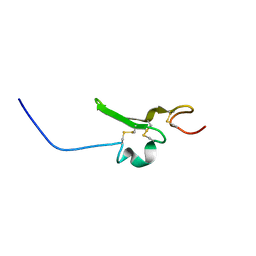 | | Solution structure of the EGF-like domain from human Amphiregulin | | Descriptor: | Amphiregulin | | Authors: | Qin, X, Hayashi, F, Terada, T, Shirouzu, M, Watanabe, S, Kigawa, T, Yabuta, N, Nojima, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the EGF-like domain from human Amphiregulin
To be Published
|
|
3EPA
 
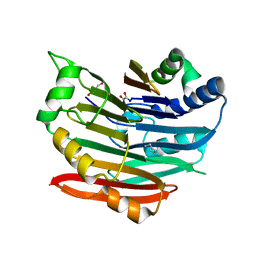 | | Human AdoMetDC E178Q mutant complexed with putrescine | | Descriptor: | 1,4-DIAMINOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRUVIC ACID, ... | | Authors: | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
2RNX
 
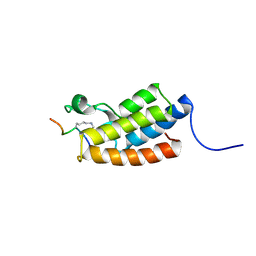 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the HUman Transcriptional Co-Activators PCAF and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
3EP3
 
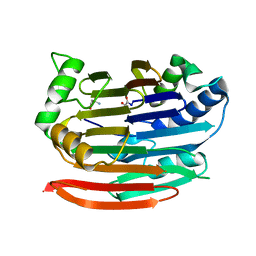 | | Human AdoMetDC D174N mutant with no putrescine bound | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRUVIC ACID, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
2RLL
 
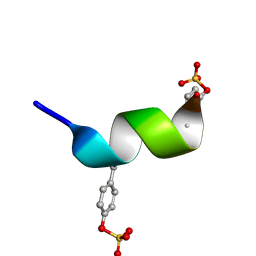 | | CCR5 Nt(7-15) | | Descriptor: | 9-mer from C-C chemokine receptor type 5 | | Authors: | Bewley, C.A, Lam, S.N. | | Deposit date: | 2007-07-21 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4
Science, 317, 2007
|
|
3IMY
 
 | |
3EXO
 
 | | Crystal structure of BACE1 bound to inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{2-methyl-5-[(6-phenylpyrimidin-4-yl)amino]phenyl}methanesulfonamide, ... | | Authors: | Allison, T.J. | | Deposit date: | 2008-10-16 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a small molecule beta-secretase inhibitor that binds without catalytic aspartate engagement.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IGG
 
 | | Novel CDK-5 inhibitors - crystal structure of inhibitor EFQ with CDK-2 | | Descriptor: | Cell division protein kinase 2, N-[1-(cis-3-hydroxycyclobutyl)-1H-imidazol-4-yl]-2-(4-methoxyphenyl)acetamide | | Authors: | Pandit, J. | | Deposit date: | 2009-07-27 | | Release date: | 2009-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and cellularly active 4-aminoimidazole inhibitors of cyclin-dependent kinase 5/p25 for the treatment of Alzheimer's disease.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2RQ8
 
 | | Solution NMR structure of titin I27 domain mutant | | Descriptor: | Titin | | Authors: | Yagawa, K, Oguro, T, Momose, T, Kawano, S, Sato, T, Endo, T. | | Deposit date: | 2009-03-05 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for unfolding pathway-dependent stability of proteins: Vectorial unfolding vs. global unfolding
Protein Sci., 2010
|
|
2RPC
 
 | |
3FFC
 
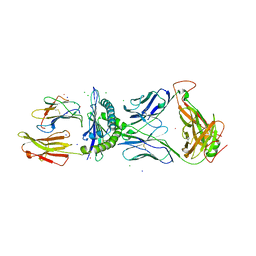 | | Crystal Structure of CF34 TCR in complex with HLA-B8/FLR | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CF34 alpha chain, ... | | Authors: | Gras, S, Burrows, S.R, Kjer-Nielsen, L, Clements, C.S, Liu, Y.C, Sullivan, L.C, Brooks, A.G, Purcell, A.W, McCluskey, J, Rossjohn, J. | | Deposit date: | 2008-12-03 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The shaping of T cell receptor recognition by self-tolerance.
Immunity, 30, 2009
|
|
3ISB
 
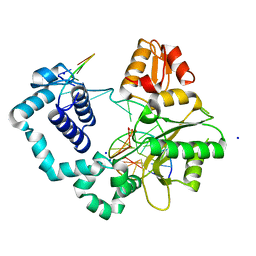 | | Binary complex of human DNA polymerase beta with a gapped DNA | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Beard, W.A, Shock, D.D, Batra, V.K, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA polymerase beta substrate specificity: side chain modulation of the "A-rule".
J.Biol.Chem., 284, 2009
|
|
2RLO
 
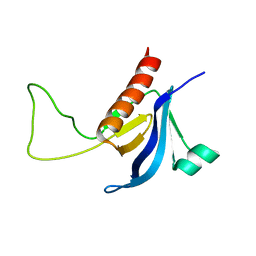 | | Split PH domain of PI3-kinase enhancer | | Descriptor: | Centaurin-gamma 1 | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2007-07-21 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Split pleckstrin homology domain-mediated cytoplasmic-nuclear localization of PI3-kinase enhancer GTPase
J.Mol.Biol., 378, 2008
|
|
3IW7
 
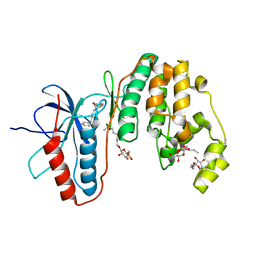 | | Human p38 MAP Kinase in Complex with an Imidazo-pyridine | | Descriptor: | 2-({4-[(4-benzylpiperidin-1-yl)carbonyl]benzyl}sulfanyl)-3H-imidazo[4,5-c]pyridine, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
2ROP
 
 | | Solution structure of domains 3 and 4 of human ATP7B | | Descriptor: | Copper-transporting ATPase 2 | | Authors: | Banci, L, Bertini, I, Cantini, F, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2008-04-04 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1
Biochemistry, 47, 2008
|
|
2RQB
 
 | | Solution structure of MDA5 CTD | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, ZINC ION | | Authors: | Takahasi, K, Kumeta, H, Tsuduki, N, Narita, R, Shigemoto, T, Hirai, R, Yoneyama, M, Horiuchi, M, Ogura, K, Fujita, T, Fuyuhiko, I. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-05 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Cytosolic RNA Sensor MDA5 and LGP2 C-terminal Domains: IDENTIFICATION OF THE RNA RECOGNITION LOOP IN RIG-I-LIKE RECEPTORS
J.Biol.Chem., 284, 2009
|
|
3I81
 
 | | Crystal structure of insulin-like growth factor 1 receptor (IGF-1R-WT) complex with BMS-754807 [1-(4-((5-cyclopropyl-1H-pyrazol-3-yl)amino)pyrrolo[2,1-f][1,2,4]triazin-2-yl)-N-(6-fluoro-3-pyridinyl)-2-methyl-L-prolinamide] | | Descriptor: | 1-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]pyrrolo[2,1-f][1,2,4]triazin-2-yl}-N-(6-fluoropyridin-3-yl)-2-methyl-L-proli namide, Insulin-like growth factor 1 receptor | | Authors: | Sack, J.S. | | Deposit date: | 2009-07-09 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of a 2,4-disubstituted pyrrolo[1,2-f][1,2,4]triazine inhibitor (BMS-754807) of insulin-like growth factor receptor (IGF-1R) kinase in clinical development.
J.Med.Chem., 52, 2009
|
|
3I8C
 
 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDR21A | | Descriptor: | DNA damage-binding protein 1, WD repeat-containing protein 21A | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-09 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3IA6
 
 | |
2RMJ
 
 | | Solution structure of RIG-I C-terminal domain | | Descriptor: | Probable ATP-dependent RNA helicase DDX58 | | Authors: | Takahasi, K, Yoneyama, M, Nihishori, T, Hirai, R, Narita, R, Gale Jr, M, Fujita, T, Inagaki, F. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nonself RNA-Sensing Mechanism of RIG-I Helicase and Activation of Antiviral Immune Responses
Mol.Cell, 29, 2008
|
|
3IE3
 
 | | Structural basis for the binding of the anti-cancer compound 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) to human glutathione S-transferases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]hexan-1-ol, GLUTATHIONE, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
