1WT7
 
 | | Solution structure of BuTX-MTX: a butantoxin-maurotoxin chimera | | Descriptor: | BuTX-MTX | | Authors: | M'Barek, S, Chagot, B, Andreotti, N, Visan, V, Mansuelle, P, Grissmer, S, Marrakchi, M, El Ayeb, M, Sampieri, F, Darbon, H, Fajloun, Z, De Waard, M, Sabatier, J.-M. | | Deposit date: | 2004-11-16 | | Release date: | 2004-11-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Increasing the molecular contacts between maurotoxin and Kv1.2 channel augments ligand affinity.
Proteins, 60, 2005
|
|
4UET
 
 | | Diversity in the structures and ligand binding sites among the fatty acid and retinol binding proteins of nematodes revealed by Na-FAR-1 from Necator americanus | | Descriptor: | NEMATODE FATTY ACID RETINOID BINDING PROTEIN | | Authors: | Rey-Burusco, M.F, Ibanez Shimabukuro, M, Griffiths, K, Cooper, A, Kennedy, M.W, Corsico, B, Smith, B.O, Griffiths, K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Diversity in the Structures and Ligand Binding Sites of Nematode Fatty Acid and Retinol Binding Proteins Revealed by Na-Far-1 from Necator Americanus.
Biochem.J., 471, 2015
|
|
4UB8
 
 | | Native structure of photosystem II (dataset-2) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
4UAC
 
 | | EUR_01830 with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Carbohydrate ABC transporter substrate-binding protein, ... | | Authors: | Koropatkin, N.M, Orlovsky, N.I. | | Deposit date: | 2014-08-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale.
Mol.Microbiol., 95, 2015
|
|
4U93
 
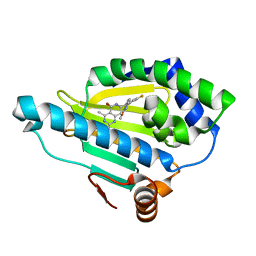 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7R)-2-amino-7-[4-fluoro-2-(6-methoxypyridin-2-yl)phenyl]-4-methyl-7,8-dihydropyrido[4,3-d]pyrimidin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
4UIO
 
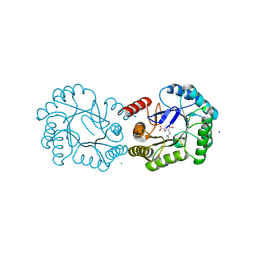 | | Structure of the Salmonella typhi Type I Dehydroquinase covalently inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (1~{R},3~{R},4~{S},5~{R})-3-methyl-1,3,4,5-tetrakis(oxidanyl)cyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Tizon, L, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Chemical Modification of a Dehydratase Enzyme Involved in Bacterial Virulence by an Ammonium Derivative: Evidence of its Active Site Covalent Adduct.
J.Am.Chem.Soc., 137, 2015
|
|
4UNE
 
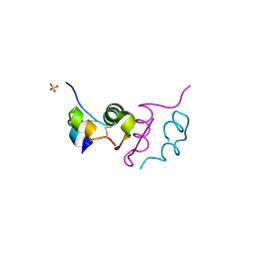 | | Human insulin B26Phe mutant crystal structure | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN, SULFATE ION | | Authors: | Zakova, L, Klevtikova, E, Lepsik, M, Collinsova, M, Watson, C.J, Turkenburg, J.P, Jiracek, J, Brzozowski, A.M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Human Insulin Analogues Modified at the B26 Site Reveal a Hormone Conformation that is Undetected in the Receptor Complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4U2U
 
 | |
4UBT
 
 | | Structure of the C93S variant of the 3-ketoacyl-CoA thiolase FadA5 from M. tuberculosis in complex with a steroid and CoA. | | Descriptor: | (2S)-2-[(8S,9S,10R,13S,14S,17R)-10,13-dimethyl-3-oxo-2,3,6,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]propanoic acid (non-preferred name), Acetyl-CoA acetyltransferase FadA5, CHLORIDE ION, ... | | Authors: | Schaefer, C.M, Kisker, C. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | FadA5 a Thiolase from Mycobacterium tuberculosis: A Steroid-Binding Pocket Reveals the Potential for Drug Development against Tuberculosis.
Structure, 23, 2015
|
|
4UDS
 
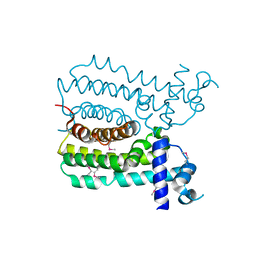 | | Crystal structure of MbdR regulator from Azoarcus sp. CIB | | Descriptor: | MBDR REGULATOR | | Authors: | Liu, H, Liu, H.X, MacMahon, S.A, Juarez, J.F, Zamarro, M.T, Eberlein, C, Boll, M, Carmona, M, Diaz, E, Naismith, J.H. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Unraveling the Specific Regulation of the Mbd Central Pathway for the Anaerobic Degradation of 3-Methylbenzoate.
J.Biol.Chem., 290, 2015
|
|
4UHV
 
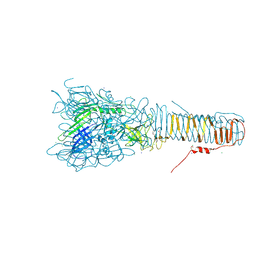 | | The structure of VgrG1, the needle tip of the bacterial Type VI Secretion System | | Descriptor: | CHLORIDE ION, SODIUM ION, VGRG1, ... | | Authors: | Spinola-Amilibia, M, Davo-Siguero, I, Ruiz, F.M, Santillana, E, Medrano, F.J, Romero, A. | | Deposit date: | 2015-03-25 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Vgrg1 from Pseudomonas Aeruginosa, the Needle Tip of the Bacterial Type Vi Secretion System
Acta Crystallogr.,Sect.D, 72, 2016
|
|
4U5P
 
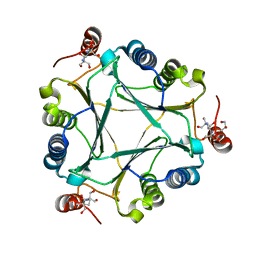 | | Crystal structure of native RhCC (YP_702633.1) from Rhodococcus jostii RHA1 at 1.78 Angstrom | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Poddar, H, Rozeboom, H.J, Thunnissen, A.M.W.H. | | Deposit date: | 2014-07-25 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Functional and structural characterization of an unusual cofactor-independent oxygenase.
Biochemistry, 54, 2015
|
|
4U5R
 
 | |
4U9B
 
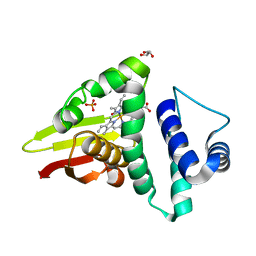 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)NO ligation state | | Descriptor: | GLYCEROL, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U99
 
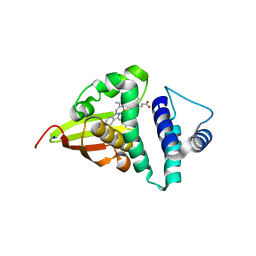 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II) ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9N
 
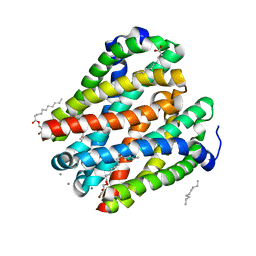 | | Structure of a membrane protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MANGANESE (II) ION, Magnesium transporter MgtE, ... | | Authors: | Takeda, H, Hattori, M, Nishizawa, T, Yamashita, K, Shah, S.T.A, Caffrey, M, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-08-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Nat Commun, 5, 2014
|
|
4U9J
 
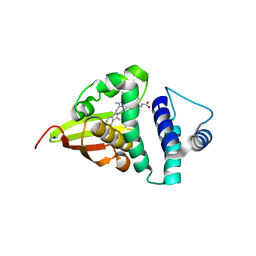 | | Crystal structure of an H-NOX protein from S. oneidensis in the Mn(II) ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, NO-binding heme-dependent sensor protein, SODIUM ION, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U96
 
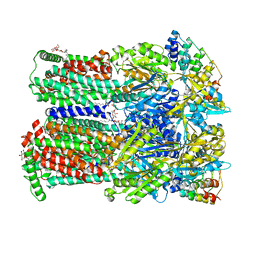 | |
4UMM
 
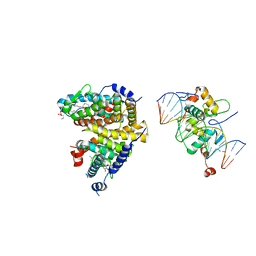 | | The Cryo-EM structure of the palindromic DNA-bound USP-EcR nuclear receptor reveals an asymmetric organization with allosteric domain positioning | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, 5'-D(*CP*AP*AP*GP*GP*GP*TP*TP*CP*AP*AP*TP*GP*CP *AP*CP*TP*TP*GP*TP)-3', 5'-D(*DGP*AP*CP*AP*AP*GP*TP*GP*CP*AP*TP*TP*GP*DAP *AP*CP*CP*CP*TP*T)-3', ... | | Authors: | Maletta, M, Orlov, I, Moras, D, Billas, I.M.L, Klaholz, B.P. | | Deposit date: | 2014-05-19 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | The Palindromic DNA-Bound Usp-Ecr Nuclear Receptor Adopts an Asymmetric Organization with Allosteric Domain Positioning.
Nat.Commun., 5, 2014
|
|
4UC4
 
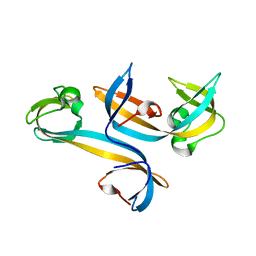 | |
4U3U
 
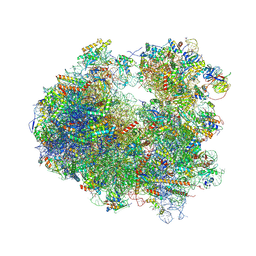 | | Crystal structure of Cycloheximide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4UF7
 
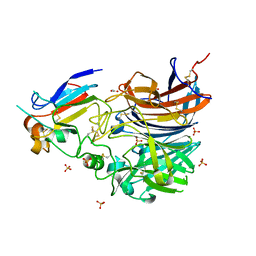 | | Ghanaian henipavirus (Gh-M74a) attachment glycoprotein in complex with human ephrinB2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lee, B, Pernet, O, Ahmed, A.A, Zeltina, A, Beaty, S.M, Bowden, T.A. | | Deposit date: | 2015-03-13 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Recognition of Human Ephrinb2 Cell Surface Receptor by an Emergent African Henipavirus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4U3L
 
 | | octameric RNA duplex co-crystallized in calcium(II)chloride | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-22 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4U6F
 
 | | Crystal structure of T-2 toxin bound to the yeast 80S ribosome | | Descriptor: | 12,13-Epoxytrichothec-9-ene-3,4,8,15-tetrol-4,15-diacetate-8-isovalerate, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-28 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U3N
 
 | | Crystal structure of CCA trinucleotide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
