6BYK
 
 | | Structure of 14-3-3 beta/alpha bound to O-ClcNAc peptide | | Descriptor: | 14-3-3 protein beta/alpha, 2-acetamido-2-deoxy-beta-D-glucopyranose, ATPPVSQASSTT O-GlcNac peptide | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
6Y6B
 
 | | Crystal structure of human 14-3-3 gamma in complex with CaMKK2 14-3-3 binding motif Ser100 and 16-OMe-Fusicoccin H | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[[(1~{S},3~{R},6~{S},7~{S},8~{R},9~{R},10~{R},11~{R},14~{S})-14-(me thoxymethyl)-3,10-dimethyl-9-oxidanyl-6-propan-2-yl-8-tricyclo[9.3.0.0^{3,7}]tetradecanyl]oxy]oxane-3,4,5-triol, 14-3-3 protein gamma, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Lentini Santo, D, Obsilova, V, Obsil, T. | | Deposit date: | 2020-02-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Stabilization of Protein-Protein Interactions between CaMKK2 and 14-3-3 by Fusicoccins.
Acs Chem.Biol., 15, 2020
|
|
4J6S
 
 | |
4ZQ0
 
 | |
8DP5
 
 | | Structure of the PEAK3/14-3-3 complex | | Descriptor: | 14-3-3 protein beta/alpha, 14-3-3 protein epsilon, Protein PEAK3, ... | | Authors: | Torosyan, H, Paul, M, Jura, N, Verba, K.A. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into regulation of the PEAK3 pseudokinase scaffold by 14-3-3.
Nat Commun, 14, 2023
|
|
5OEG
 
 | | Molecular tweezers modulate 14-3-3 protein-protein interactions | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-3,22-Bis(dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaene, 14-3-3 protein sigma, CHLORIDE ION | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2017-07-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Molecular tweezers modulate 14-3-3 protein-protein interactions.
Nat Chem, 5, 2013
|
|
7C8E
 
 | | Crystal Structure of 14-3-3 epsilon with 9J10 peptide | | Descriptor: | 14-3-3 protein epsilon, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9J10 | | Authors: | Mathivanan, S, Sudhakar, S, Bairy, S, Kamariah, N, Venkitaraman, A. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Target identification for small-molecule discovery in the FOXO3a tumor-suppressor pathway using a biodiverse peptide library.
Cell Chem Biol, 28, 2021
|
|
8DGN
 
 | |
4F7R
 
 | |
8DGM
 
 | | 14-3-3 epsilon bound to phosphorylated PEAK1 (pT1165) peptide | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, Inactive tyrosine-protein kinase PEAK1 | | Authors: | Roy, M.J, Hardy, J.M, Lucet, I.S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mapping of PEAK pseudokinase interactions identifies 14-3-3 as a molecular switch for PEAK3 signaling.
Nat Commun, 14, 2023
|
|
5OKF
 
 | | CH1 chimera of human 14-3-3 sigma with the HSPB6 phosphopeptide in a conformation with self-bound phosphopeptides | | Descriptor: | 14-3-3 protein sigma,Heat shock protein beta-6, CADMIUM ION | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chimeric 14-3-3 proteins for unraveling interactions with intrinsically disordered partners.
Sci Rep, 7, 2017
|
|
5OM0
 
 | | CH2 chimera of human 14-3-3 sigma with the Gli1 phosphopeptide around Ser640 | | Descriptor: | 14-3-3 protein sigma,Zinc finger protein GLI1, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chimeric 14-3-3 proteins for unraveling interactions with intrinsically disordered partners.
Sci Rep, 7, 2017
|
|
8Q1S
 
 | | Pathogenic mutations of human phosphorylation sites affect protein-protein interactions | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, BROMIDE ION, ... | | Authors: | Roske, Y, Daumke, O, Rrustemi, T, Selbach, M. | | Deposit date: | 2023-08-01 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Pathogenic mutations of human phosphorylation sites affect protein-protein interactions.
Nat Commun, 15, 2024
|
|
6GNJ
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form in complex with STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GN0
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, tetrameric crystal form | | Descriptor: | 14-3-3 protein beta/alpha, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-29 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
3M51
 
 | | Structure of the 14-3-3/PMA2 complex stabilized by Pyrrolidone1 | | Descriptor: | 14-3-3-like protein C, 2-hydroxy-5-[(5S)-3-hydroxy-5-(4-nitrophenyl)-2-oxo-4-(phenylcarbonyl)-2,5-dihydro-1H-pyrrol-1-yl]benzoic acid, N.plumbaginifolia H+-translocating ATPase mRNA | | Authors: | Ottmann, C, Rose, R, Waldmann, H. | | Deposit date: | 2010-03-12 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Identification and structure of small-molecule stabilizers of 14-3-3 protein-protein interactions
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
5NWK
 
 | | 14-3-3c in complex with CPP and fusicoccin | | Descriptor: | 14-3-3 c-1 protein, FUSICOCCIN, PHOSPHOSERINE, ... | | Authors: | Saponaro, A, Porro, A, Chaves-Sanjuan, A, Nardini, M, Thiel, G, Moroni, A. | | Deposit date: | 2017-05-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fusicoccin Activates KAT1 Channels by Stabilizing Their Interaction with 14-3-3 Proteins.
Plant Cell, 29, 2017
|
|
6XAG
 
 | | Apo BRAF dimer bound to 14-3-3 | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Liau, N.P.D, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2020-06-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dimerization Induced by C-Terminal 14-3-3 Binding Is Sufficient for BRAF Kinase Activation.
Biochemistry, 59, 2020
|
|
1A38
 
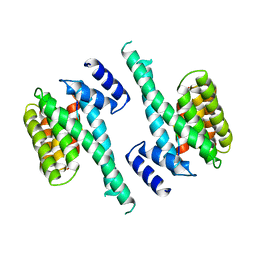 | | 14-3-3 PROTEIN ZETA BOUND TO R18 PEPTIDE | | Descriptor: | 14-3-3 PROTEIN ZETA, R18 PEPTIDE (PHCVPRDLSWLDLEANMCLP) | | Authors: | Petosa, C, Masters, S.C, Pohl, J, Wang, B, Fu, H, Liddington, R.C. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove.
J.Biol.Chem., 273, 1998
|
|
6BYL
 
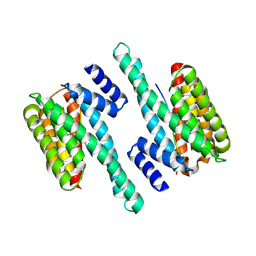 | | Structure of 14-3-3 gamma bound to O-GlcNAcylated thr peptide | | Descriptor: | 14-3-3 protein gamma, 2-acetamido-2-deoxy-beta-D-glucopyranose, TSASTTVPVTTATTTTTSTW O-GlcNac peptide | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
4DX0
 
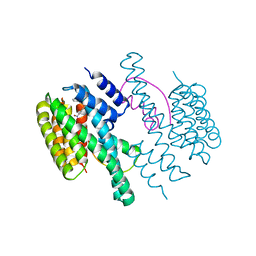 | | Structure of the 14-3-3/PMA2 complex stabilized by a pyrazole derivative | | Descriptor: | 14-3-3-like protein E, 4-[(4R)-4-(4-nitrophenyl)-6-oxidanylidene-3-phenyl-1,4-dihydropyrrolo[3,4-c]pyrazol-5-yl]benzoic acid, N.plumbaginifolia H+-translocating ATPase mRNA | | Authors: | Richter, A, Rose, R, Hedberg, C, Waldmann, H, Ottmann, C. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Optimised Small-Molecule Stabiliser of the 14-3-3-PMA2 Protein-Protein Interaction.
Chemistry, 18, 2012
|
|
8CPD
 
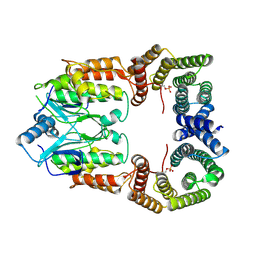 | | Cryo-EM structure of CRaf dimer with 14:3:3 | | Descriptor: | 14-3-3 protein zeta isoform X1, RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Dedden, D, Graedler, U, Schwarz, D, Thomsen, M, Leuthner, B, Schneider, E, Nitsche, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Cryo-EM Structures of CRAF 2 /14-3-3 2 and CRAF 2 /14-3-3 2 /MEK1 2 Complexes.
J.Mol.Biol., 436, 2024
|
|
8QTC
 
 | |
1A37
 
 | | 14-3-3 PROTEIN ZETA BOUND TO PS-RAF259 PEPTIDE | | Descriptor: | 14-3-3 PROTEIN ZETA, PS-RAF259 PEPTIDE LSQRQRST(SEP)TPNVHM | | Authors: | Petosa, C, Masters, S.C, Pohl, J, Wang, B, Fu, H, Liddington, R.C. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove.
J.Biol.Chem., 273, 1998
|
|
7MFD
 
 | | Autoinhibited BRAF:(14-3-3)2:MEK complex with the BRAF RBD resolved | | Descriptor: | 14-3-3 protein zeta/delta, Dual specificity mitogen-activated protein kinase kinase 1, N-(3-fluoro-4-{[4-methyl-2-oxo-7-(pyrimidin-2-yloxy)-2H-chromen-3-yl]methyl}pyridin-2-yl)-N'-methylsulfuric diamide, ... | | Authors: | Martinez Fiesco, J.A, Ping, Z, Durrant, D.E, Morrison, D.K. | | Deposit date: | 2021-04-09 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the BRAF monomer-to-dimer transition mediated by RAS binding.
Nat Commun, 13, 2022
|
|
