8SO0
 
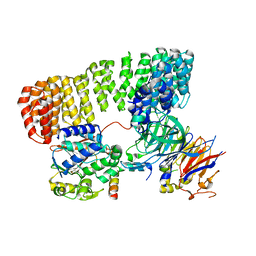 | | Cryo-EM structure of the PP2A:B55-FAM122A complex | | Descriptor: | FE (III) ION, PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-04-28 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.79961 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
8BRN
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with an alpha-aminonaphthylmethylphosphonic acid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Hussein, W.M, Guddat, L.W, Schenk, G. | | Deposit date: | 2022-11-23 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of an alpha-aminonaphthylmethylphosphonic acid inhibitor of purple acid phosphatase using rational structure-based design approaches.
Eur.J.Med.Chem., 254, 2023
|
|
8PQ7
 
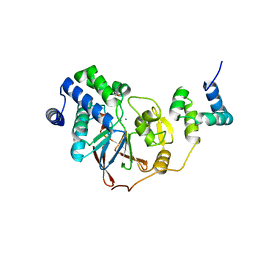 | |
8B5R
 
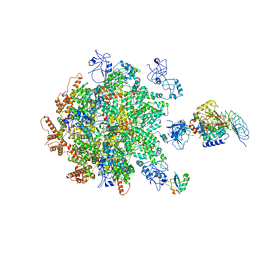 | | p97-p37-SPI substrate complex | | Descriptor: | I3 sequence being threaded through the p97 channel, Protein phosphatase 1 regulatory subunit 7, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit, ... | | Authors: | van den Boom, J, Marini, G, Meyer, H, Saibil, H. | | Deposit date: | 2022-09-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural basis of ubiquitin-independent PP1 complex disassembly by p97.
Embo J., 42, 2023
|
|
8GFT
 
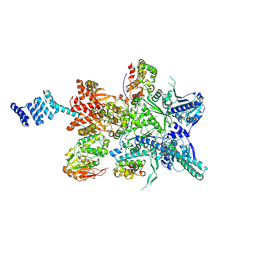 | | Hsp90 provides platform for CRaf dephosphorylation by PP5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, ... | | Authors: | Jaime-Garza, M, Nowotny, C.A, Coutandin, D, Wang, F, Tabios, M, Agard, D.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Hsp90 provides a platform for kinase dephosphorylation by PP5.
Nat Commun, 14, 2023
|
|
8GAE
 
 | | Hsp90 provides platform for CRaf dephosphorylation by PP5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, ... | | Authors: | Jaime-Garza, M, Nowotny, C.A, Coutandin, D, Wang, F, Tabios, M, Agard, D.A. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Hsp90 provides a platform for kinase dephosphorylation by PP5.
Nat Commun, 14, 2023
|
|
7SS6
 
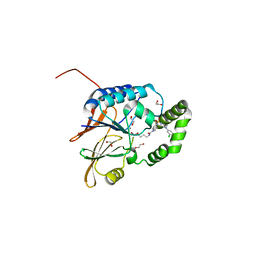 | | Structure of Klebsiella LpxH in complex with JH-LPH-45 | | Descriptor: | 1,2-ETHANEDIOL, 5-{4-[3-chloro-5-(trifluoromethyl)phenyl]piperazine-1-sulfonyl}-N-[5-(hydroxyamino)-5-oxopentyl]-2,3-dihydro-1H-indole-1-carboxamide, MANGANESE (II) ION, ... | | Authors: | Cho, J, Cochrane, C.S, Zhou, P. | | Deposit date: | 2021-11-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of LpxH Inhibitors Chelating the Active Site Dimanganese Metal Cluster of LpxH.
Chemmedchem, 18, 2023
|
|
7SS7
 
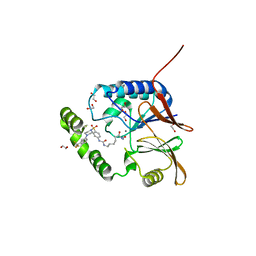 | | Crystal structure of Klebsiella LpxH in complex with JH-LPH-50 | | Descriptor: | 1,2-ETHANEDIOL, 6-{[(4-{4-[3-chloro-5-(trifluoromethyl)phenyl]piperazine-1-sulfonyl}phenyl)carbamoyl]amino}-N-hydroxyhexanamide, MANGANESE (II) ION, ... | | Authors: | Cho, J, Cochrane, C.S, Zhou, P. | | Deposit date: | 2021-11-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Development of LpxH Inhibitors Chelating the Active Site Dimanganese Metal Cluster of LpxH.
Chemmedchem, 18, 2023
|
|
7YCX
 
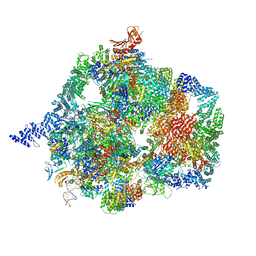 | | The structure of INTAC-PEC complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1,DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Zheng, H, Jin, Q, Wang, X, Qi, Y, Liu, W, Ren, Y, Zhao, D, Chen, F.X, Cheng, J, Chen, X, Xu, Y. | | Deposit date: | 2022-07-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural basis of INTAC-regulated transcription.
Protein Cell, 14, 2023
|
|
8DWL
 
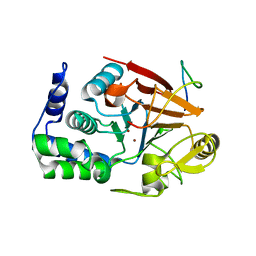 | | Inhibitor-3:PP1 coexpressed complex | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase PPP1R11, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
8DWK
 
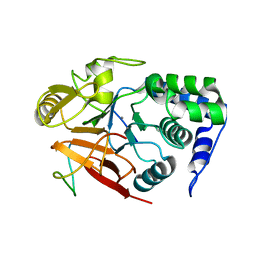 | | Inhibitor-3:PP1 reconstituted complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
7QWJ
 
 | |
7ZR1
 
 | | Chaetomium thermophilum Mre11-Rad50-Nbs1 complex bound to ATPyS (composite structure) | | Descriptor: | DH domain-containing protein, Double-strand break repair protein, FHA domain-containing protein, ... | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-05-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
8BAH
 
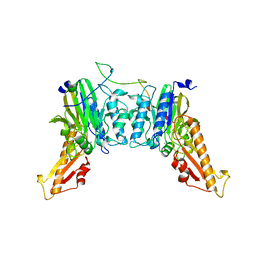 | | Human Mre11-Nbs1 complex | | Descriptor: | Double-strand break repair protein MRE11, MANGANESE (II) ION, Nibrin | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-10-11 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
7ZR6
 
 | |
7ZR5
 
 | |
7T0Y
 
 | | The Ribosomal RNA Processing 1B Protein Phosphatase-1 Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, FLUORIDE ION, ... | | Authors: | Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The ribosomal RNA processing 1B:protein phosphatase 1 holoenzyme reveals non-canonical PP1 interaction motifs.
Cell Rep, 41, 2022
|
|
7QFB
 
 | | Crystal structure of Protein Phosphatase 1 in complex with PP1-binding peptide from PTG | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Protein phosphatase 1 regulatory subunit 3C, ... | | Authors: | Semrau, M.S, Storici, P, Lolli, G. | | Deposit date: | 2021-12-05 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular architecture of the glycogen- committed PP1/PTG holoenzyme.
Nat Commun, 13, 2022
|
|
7QM2
 
 | |
7SOY
 
 | | The structure of the PP2A-B56gamma1 holoenzyme-PME-1 complex | | Descriptor: | Isoform Gamma-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, Protein phosphatase methylesterase 1, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Li, Y, Balakrishnan, V.K, Rowse, M, Novikova, I.V, Xing, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Coupling to short linear motifs creates versatile PME-1 activities in PP2A holoenzyme demethylation and inhibition.
Elife, 11, 2022
|
|
8DZK
 
 | | Dbr1 in complex with 5-mer cleavage product | | Descriptor: | FE (II) ION, RNA (5'-R(P*(G46)P*UP*GP*UP*U)-3'), RNA lariat debranching enzyme, ... | | Authors: | Clark, N.E, Taylor, A.B. | | Deposit date: | 2022-08-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the RNA Lariat Debranching Enzyme Dbr1 with Hydrolyzed Phosphorothioate RNA Product.
Biochemistry, 61, 2022
|
|
7YZP
 
 | | Hairpin-bound state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and a DNA hairpin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA hairpin (59-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7YZO
 
 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7Z03
 
 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and extended dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (39-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7TXH
 
 | | Human MRas Q71R in complex with human Shoc2 LRR domain M173I and human PP1Ca | | Descriptor: | GLYCEROL, Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, ... | | Authors: | Hauseman, Z.J, Viscomi, J, Dhembi, A, Clark, K, King, D.A, Fodor, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the MRAS-SHOC2-PP1C phosphatase complex.
Nature, 609, 2022
|
|
