7ULS
 
 | | Recombinant muscarinic toxin alpha | | Descriptor: | GLYCEROL, Muscarinic toxin alpha | | Authors: | Xu, J, Lei, X, Chen, L. | | Deposit date: | 2022-04-05 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | recombinant MTalpha at 1.8 Angstroms resolution
To Be Published
|
|
3W12
 
 | | Insulin receptor ectodomain construct comprising domains L1-CR in complex with high-affinity insulin analogue [D-PRO-B26]-DTI-NH2, alpha-CT peptide(704-719) and FAB 83-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | Authors: | Lawrence, M.C, Smith, B.J, Brzozowsk, A.M. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.301 Å) | | Cite: | How insulin engages its primary binding site on the insulin receptor
Nature, 493, 2013
|
|
4GNE
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-7 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GNG
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3K9me3 peptide | | Descriptor: | GLYCEROL, Histone H3.3, Histone-lysine N-methyltransferase NSD3, ... | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
5MUI
 
 | | Glycoside hydrolase BT_0996 | | Descriptor: | Beta-galactosidase, beta-L-arabinofuranose-(1-2)-alpha-L-rhamnopyranose-(1-2)-[alpha-L-rhamnopyranose-(1-3)]alpha-L-arabinopyranose-(1-4)-[4-O-[(1R)-1-hydroxyethyl]-2-O-methyl-alpha-L-fucopyranose-(1-2)]beta-D-galactopyranose-(1-2)-alpha-D-aceric acid-(1-3)-alpha-L-rhamnopyranose | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
4ZOK
 
 | |
9MIN
 
 | |
5MWK
 
 | | Glycoside hydrolase BT_0986 | | Descriptor: | BROMIDE ION, CALCIUM ION, Glycoside hydrolase family 2, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
6I0X
 
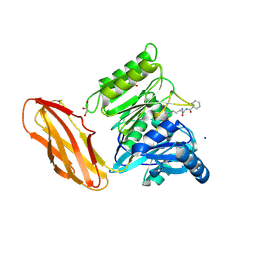 | | Porphyromonas gingivalis peptidylarginine deminase (PPAD) mutant G231N/E232T/N235D in complex with Cl-amidine. | | Descriptor: | GLYCEROL, N-[(1S)-1-(AMINOCARBONYL)-4-(ETHANIMIDOYLAMINO)BUTYL]BENZAMIDE, Peptidylarginine deiminase, ... | | Authors: | Gomis-Ruth, F.X, Goulas, T, Sola, M, Potempa, J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure, function, and inhibition of a genomic/clinical variant of Porphyromonas gingivalis peptidylarginine deiminase.
Protein Sci., 28, 2019
|
|
6UXW
 
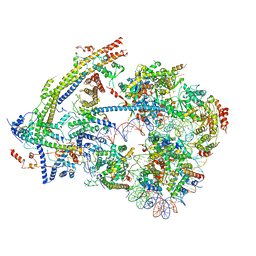 | | SWI/SNF nucleosome complex with ADP-BeFx | | Descriptor: | 601 sequence bottom strand, 601 sequence top strand, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | He, Y, Han, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (8.96 Å) | | Cite: | Cryo-EM structure of SWI/SNF complex bound to a nucleosome.
Nature, 579, 2020
|
|
4ZQD
 
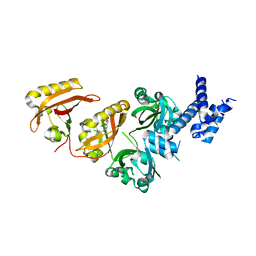 | | Crystal Structure of the Heterodimeric HIF-2a:ARNT Complex with the Benzoxadiazole Antagonist 0X3 | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(3-chloro-5-fluorophenyl)-4-nitro-2,1,3-benzoxadiazol-5-amine | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-09 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
7PSA
 
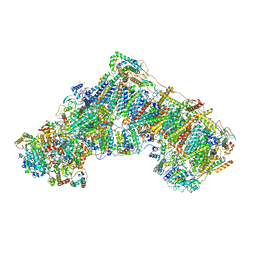 | | The acetogenin-bound complex I of Mus musculus resolved to 3.4 angstroms | | Descriptor: | (3~{S},5~{S})-5-methyl-3-[(13~{R})-13-oxidanyl-13-[(2~{R},5~{R})-5-[(2~{R},5~{R})-5-[(1~{R})-1-oxidanylundecyl]oxolan-2-yl]oxolan-2-yl]tridecyl]oxolan-2-one, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Grba, D, Hirst, J. | | Deposit date: | 2021-09-22 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-electron microscopy reveals how acetogenins inhibit mitochondrial respiratory complex I.
J.Biol.Chem., 298, 2022
|
|
1X1J
 
 | | Crystal Structure of Xanthan Lyase (N194A) with a Substrate. | | Descriptor: | (4AR,6R,7S,8R,8AR)-8-((5R,6R)-3-CARBOXY-TETRAHYDRO-4,5,6-TRIHYDROXY-2H-PYRAN-2-YLOXY)-HEXAHYDRO-6,7-DIHYDROXY-2-METHYLPYRANO[3,2-D][1,3]DIOXINE-2-CARBOXYLIC ACID), CALCIUM ION, xanthan lyase | | Authors: | Maruyama, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2005-04-04 | | Release date: | 2005-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase Complexed with a Substrate: Insights into the Enzyme Reaction Mechanism
J.Mol.Biol., 350, 2005
|
|
9EW1
 
 | | Ternary structure of 14-3-3s, CRAF phosphopeptide (pS259) and compound 79 (1124379). | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-1-[8-(4-iodophenyl)sulfonyl-5-oxa-2,8-diazaspiro[3.5]nonan-2-yl]ethanone, CHLORIDE ION, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2024-04-03 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
6I5G
 
 | | X-ray structure of human soluble Epoxide Hydrolase C-terminal Domain (hsEH CTD)in complex with 15d-PGJ2 | | Descriptor: | (5E,14E)-11-oxoprosta-5,9,12,14-tetraen-1-oic acid, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | Authors: | Abis, G, Kopec, J, Yue, W.W, Conte, M.R. | | Deposit date: | 2018-11-13 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 15-deoxy-Delta12,14-Prostaglandin J2inhibits human soluble epoxide hydrolase by a dual orthosteric and allosteric mechanism.
Commun Biol, 2, 2019
|
|
4U79
 
 | | Crystal structure of human JNK3 in complex with a benzenesulfonamide inhibitor. | | Descriptor: | Mitogen-activated protein kinase 10, N-{4-[(3-{2-[(trans-4-aminocyclohexyl)amino]pyrimidin-4-yl}pyridin-2-yl)oxy]naphthalen-1-yl}benzenesulfonamide | | Authors: | Mohr, C. | | Deposit date: | 2014-07-30 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Unfolded Protein Response in Cancer: IRE1 alpha Inhibition by Selective Kinase Ligands Does Not Impair Tumor Cell Viability.
Acs Med.Chem.Lett., 6, 2015
|
|
7L61
 
 | |
6US2
 
 | | MTH1 in complex with compound 5 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N-[5-(2,3-dimethylphenyl)-1,2,3,4-tetrahydro-1,6-naphthyridin-7-yl]acetamide | | Authors: | Newby, Z.E.R, Lansdon, E.B. | | Deposit date: | 2019-10-24 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.80012655 Å) | | Cite: | Discovery of Potent and Selective MTH1 Inhibitors for Oncology: Enabling Rapid Target (In)Validation.
Acs Med.Chem.Lett., 11, 2020
|
|
7VUX
 
 | | Complex structure of PD1 and 609A-Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
6UWP
 
 | | BACE-1 in complex with compound #32 | | Descriptor: | (1R,2R)-2-[(4aR,7aR)-2-amino-6-(pyrimidin-2-yl)-4a,5,6,7-tetrahydropyrrolo[3,4-d][1,3]thiazin-7a(4H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
9F0R
 
 | | VIM-2 in complex with GKV65 (5g) - dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors | | Descriptor: | FORMIC ACID, MAGNESIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Bosman, R, Prester, A, Bartels, K, Gulyas, K.V, Erdelyi, M, Schulz, E.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors.
Commun Chem, 8, 2025
|
|
4GVQ
 
 | | X-ray structure of the Archaeoglobus fulgidus methenyl-tetrahydromethanopterin cyclohydrolase in complex with tetrahydromethanpterin | | Descriptor: | 1-[4-({(1R)-1-[(6S,7S)-2-amino-7-methyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]ethyl}amino)phenyl]-1-deoxy-5-O-{5-O-[(R)-{[(1R)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-xylitol, Methenyltetrahydromethanopterin cyclohydrolase | | Authors: | Upadhyay, V, Demmer, U, Warkentin, E, Moll, J, Shima, S, Ermler, U. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and catalytic mechanism of N(5),N(10)-methenyl-tetrahydromethanopterin cyclohydrolase.
Biochemistry, 51, 2012
|
|
6I75
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative 2 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-4-[4-[2,3,5,6-tetrakis(fluoranyl)-4-oxidanyl-phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.171 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
6UY0
 
 | |
5MZO
 
 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (open conformation) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
