5E1A
 
 | | Structure of KcsA with L24C/R117C mutations | | Descriptor: | POTASSIUM ION, antibody Fab fragment heavy chain, antibody Fab fragment light chain, ... | | Authors: | Upadhyay, V, Kim, D.M, Nimigean, C.M. | | Deposit date: | 2015-09-29 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Conformational heterogeneity in closed and open states of the KcsA potassium channel in lipid bicelles.
J.Gen.Physiol., 148, 2016
|
|
4GVR
 
 | | X-ray structure of the Archaeoglobus fulgidus methenyl-tetrahydromethanopterin cyclohydrolase | | Descriptor: | Methenyltetrahydromethanopterin cyclohydrolase | | Authors: | Upadhyay, V, Demmer, U, Warkentin, E, Moll, J, Shima, S, Ermler, U. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure and catalytic mechanism of N(5),N(10)-methenyl-tetrahydromethanopterin cyclohydrolase.
Biochemistry, 51, 2012
|
|
4GVQ
 
 | | X-ray structure of the Archaeoglobus fulgidus methenyl-tetrahydromethanopterin cyclohydrolase in complex with tetrahydromethanpterin | | Descriptor: | 1-[4-({(1R)-1-[(6S,7S)-2-amino-7-methyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]ethyl}amino)phenyl]-1-deoxy-5-O-{5-O-[(R)-{[(1R)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-xylitol, Methenyltetrahydromethanopterin cyclohydrolase | | Authors: | Upadhyay, V, Demmer, U, Warkentin, E, Moll, J, Shima, S, Ermler, U. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and catalytic mechanism of N(5),N(10)-methenyl-tetrahydromethanopterin cyclohydrolase.
Biochemistry, 51, 2012
|
|
4GVS
 
 | | X-ray structure of the Archaeoglobus fulgidus methenyl-tetrahydromethanopterin cyclohydrolase in complex with N5-formyl-tetrahydromethanopterin | | Descriptor: | 1-[4-({(1R)-1-[(6S,7R)-2-amino-5-formyl-7-methyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]ethyl}amino)phenyl]-1-deoxy-5 -O-{5-O-[(R)-{[(1R)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-ribitol, Methenyltetrahydromethanopterin cyclohydrolase | | Authors: | Upadhyay, V, Demmer, U, Warkentin, E, Moll, J, Shima, S, Ermler, U. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and catalytic mechanism of N(5),N(10)-methenyl-tetrahydromethanopterin cyclohydrolase.
Biochemistry, 51, 2012
|
|
4KPU
 
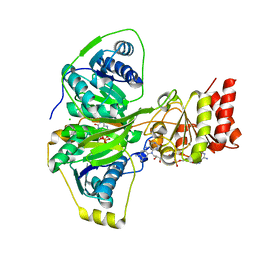 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | CHLORIDE ION, Electron transfer flavoprotein alpha subunit, Electron transfer flavoprotein alpha/beta-subunit, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-05-14 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
4L1F
 
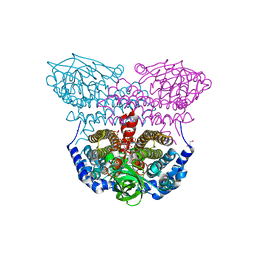 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | 1,3-PROPANDIOL, Acyl-CoA dehydrogenase domain protein, COENZYME A PERSULFIDE, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-06-03 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
6EO1
 
 | | The electron crystallography structure of the cAMP-bound potassium channel MloK1 (PCO-refined) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Kowal, J, Biyani, N, Chami, M, Scherer, S, Rzepiela, A, Baumgartner, P, Upadhyay, V, Nimigean, C, Stahlberg, H. | | Deposit date: | 2017-10-08 | | Release date: | 2017-12-27 | | Last modified: | 2018-01-10 | | Method: | ELECTRON CRYSTALLOGRAPHY (4.5 Å) | | Cite: | High-Resolution Cryoelectron Microscopy Structure of the Cyclic Nucleotide-Modulated Potassium Channel MloK1 in a Lipid Bilayer.
Structure, 26, 2018
|
|
4L2I
 
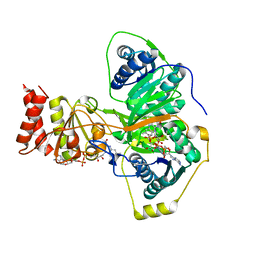 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | CHLORIDE ION, Electron transfer flavoprotein alpha subunit, Electron transfer flavoprotein alpha/beta-subunit, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-06-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
