8EUU
 
 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
8EUW
 
 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
7SR6
 
 | | Human Endogenous Retrovirus (HERV-K) reverse transcriptase ternary complex with dsDNA template Primer and dNTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Baldwin, E.T, Nichols, C. | | Deposit date: | 2021-11-08 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Human endogenous retrovirus-K (HERV-K) reverse transcriptase (RT) structure and biochemistry reveals remarkable similarities to HIV-1 RT and opportunities for HERV-K-specific inhibition.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8TD0
 
 | | Structure of PYCR1 complexed with 5-oxo-7a-phenyl-hexahydropyrrolo[2,1-b][1,3]thiazole-3-carboxylic acid | | Descriptor: | (3R,4S,7aR)-5-oxo-7a-phenylhexahydropyrrolo[2,1-b][1,3]thiazole-3-carboxylic acid, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
5J7F
 
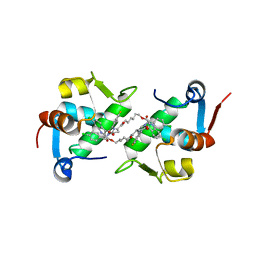 | | Structure of MDM2 with low molecular weight inhibitor with aliphatic linker. | | Descriptor: | 4-({6-[(6-chloro-3-{1-[(4-chlorophenyl)methyl]-4-(4-fluorophenyl)-1H-imidazol-5-yl}-1H-indole-2-carbonyl)oxy]hexyl}amino)-4-oxobutanoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Kubica, K, Guzik, K, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4,5-Trisubstituted Imidazole-Based p53-MDM2/MDMX Antagonists with Aliphatic Linkers for Conjugation with Biological Carriers.
J. Med. Chem., 60, 2017
|
|
7ZWG
 
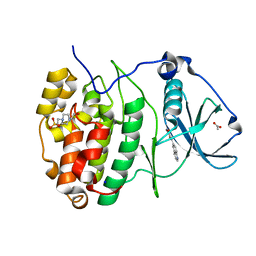 | | The Crystal structure of RO4493940 bound to CK2alpha | | Descriptor: | (5~{Z})-5-(quinolin-6-ylmethylidene)-1,3-thiazolidine-2,4-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Chemical proteomics reveals the target landscape of 1,000 kinase inhibitors.
Nat.Chem.Biol., 20, 2024
|
|
7ZWE
 
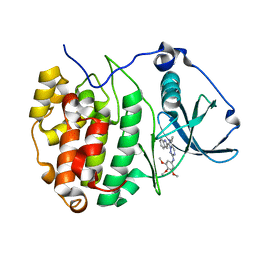 | | The Crystal structure of GW8695 bound to CK2alpha | | Descriptor: | 7-(1~{H}-indol-2-yl)-5-methyl-~{N}-(3,4,5-trimethoxyphenyl)imidazo[5,1-f][1,2,4]triazin-2-amine, Casein kinase II subunit alpha | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Chemical proteomics reveals the target landscape of 1,000 kinase inhibitors.
Nat.Chem.Biol., 20, 2024
|
|
7A4Q
 
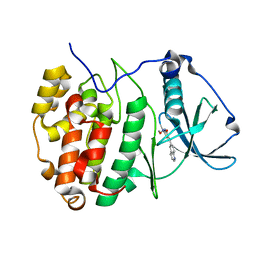 | | The Crystal structure of RO4613269 bound to CK2alpha | | Descriptor: | 2-methoxyimino-5-(quinolin-6-ylmethyl)-1,3-thiazol-4-one, Casein kinase II subunit alpha | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2020-08-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Chemical proteomics reveals the target landscape of 1,000 kinase inhibitors.
Nat.Chem.Biol., 2023
|
|
5J7G
 
 | | Structure of MDM2 with low molecular weight inhibitor with aliphatic linker. | | Descriptor: | 4-({6-[(6-chloro-3-{1-[(4-chlorophenyl)methyl]-4-(4-fluorophenyl)-1H-imidazol-5-yl}-1H-indole-2-carbonyl)oxy]hexyl}amino)-4-oxobutanoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Kubica, K, Guzik, K, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,4,5-Trisubstituted Imidazole-Based p53-MDM2/MDMX Antagonists with Aliphatic Linkers for Conjugation with Biological Carriers.
J. Med. Chem., 60, 2017
|
|
6VQN
 
 | | Co-crystal structure of human PD-L1 complexed with Compound A | | Descriptor: | N,N'-(2,2'-dimethyl[1,1'-biphenyl]-3,3'-diyl)bis(5-{[(2-hydroxyethyl)amino]methyl}pyridine-2-carboxamide), Programmed cell death 1 ligand 1 | | Authors: | White, A, Lakshminarasimhan, D, Leo, C, Suto, R.K. | | Deposit date: | 2020-02-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Checkpoint inhibition through small molecule-induced internalization of programmed death-ligand 1.
Nat Commun, 12, 2021
|
|
6DFQ
 
 | | mouse diabetogenic TCR I.29 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, TCR alpha chain, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2018-05-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes.
Sci Immunol, 4, 2019
|
|
9FKZ
 
 | | Discovery of a Series of Covalent, Cell Active Bfl-1 Inhibitors | | Descriptor: | Bcl-2-related protein A1, ~{N}-[4-[(1~{R},3~{R})-3-azanylcyclopentyl]oxyphenyl]-~{N}-[(4-chlorophenyl)methyl]prop-2-enamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
J.Med.Chem., 67, 2024
|
|
8U31
 
 | | Crystal structure of PD-1 in complex with a Fab | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Sun, D, Masureel, M. | | Deposit date: | 2023-09-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure- and machine learning-guided engineering demonstrate that a non-canonical disulfide in an anti-PD-1 rabbit antibody does not impede antibody developability.
Mabs, 16, 2024
|
|
5WVD
 
 | | Structure of Mnk1 in complex with DS12881479 | | Descriptor: | 1-methyl-N-(5-phenyl-1,3-thiazol-2-yl)piperidine-4-carboxamide, MAP kinase interacting serine/threonine kinase 1, SULFATE ION | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2016-12-24 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel inhibitor stabilizes the inactive conformation of MAPK-interacting kinase 1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6VUB
 
 | |
9FZA
 
 | |
6VUF
 
 | |
7TM5
 
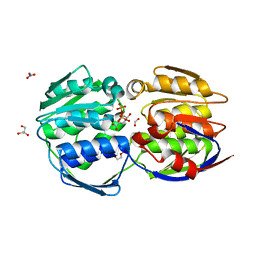 | |
6VWI
 
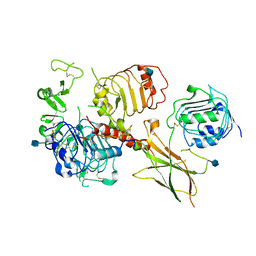 | | Head region of the closed conformation of the human type 1 insulin-like growth factor receptor ectodomain in complex with human insulin-like growth factor II. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin-like growth factor II, Leucine-zippered human type 1 insulin-like growth factor receptor ectodomain | | Authors: | Xu, Y, Kirk, N.S, Lawrence, M.C, Croll, T.I. | | Deposit date: | 2020-02-19 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | How IGF-II Binds to the Human Type 1 Insulin-like Growth Factor Receptor.
Structure, 28, 2020
|
|
8TZI
 
 | | Human equilibrative nucleoside transporter-1, JH-ENT-01 bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[4-[3-[3,4-dimethoxy-5-[[4-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]methoxy]phenyl]carbonyloxypropyl]-1,4-diazepan-1-yl]propyl 3,4,5-trimethoxybenzoate, Equilibrative nucleoside transporter 1 | | Authors: | Wright, N.J, Lee, S.Y. | | Deposit date: | 2023-08-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of an equilibrative nucleoside transporter subtype 1 inhibitor for pain relief.
Nat Commun, 15, 2024
|
|
6W4M
 
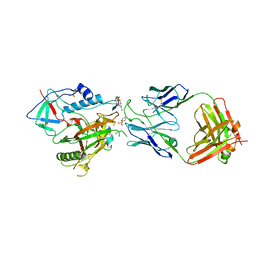 | | CRYSTAL STRUCTURE OF THE ADCC-POTENT, WEAKLY NEUTRALIZING HIV ENV CO-RECEPTOR BINDING SITE ANTIBODY N12-I2 FAB IN COMPLEX WITH HIV-1 CLADE A/E GP120 AND M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTI-HIV ANTIBODY N12-I2 FAB HEAVY CHAIN, ANTI-HIV ANTIBODY N12-I2 FAB LIGHT CHAIN, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Defining rules governing recognition and Fc-mediated effector functions to the HIV-1 co-receptor binding site.
Bmc Biol., 18, 2020
|
|
6VKD
 
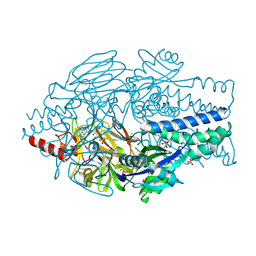 | | Crystal Structure of Inhibitor JNJ-36689282 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 1-cyclopropyl-3-({1-[3-(methylsulfonyl)propyl]-1H-pyrrolo[3,2-c]pyridin-2-yl}methyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one, CHLORIDE ION, Prefusion RSV F (DS-Cav1), ... | | Authors: | McLellan, J.S. | | Deposit date: | 2020-01-20 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of 3-({5-Chloro-1-[3-(methylsulfonyl)propyl]-1H-indol-2-yl}methyl)-1-(2,2,2-trifluoroethyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one (JNJ-53718678), a Potent and Orally Bioavailable Fusion Inhibitor of Respiratory Syncytial Virus.
J.Med.Chem., 63, 2020
|
|
8C63
 
 | | Crystal structure of arsenoplatin-1/B-DNA adduct obtained upon 4 h of soaking | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, PLATINUM (II) ION, ... | | Authors: | Troisi, R, Tito, G, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
8AYS
 
 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 4-(2-aminothiazol-4-yl)phenol | | Descriptor: | 4-(2-amino-1,3-thiazol-4-yl)phenol, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
8C64
 
 | | Crystal structure of arsenoplatin-1/B-DNA adduct obtained upon 48 h of soaking | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PLATINUM (II) ION, arsenoplatin-1 | | Authors: | Troisi, R, Tito, G, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
