4ZTD
 
 | | Crystal Structure of Human PCNA in complex with a TRAIP peptide | | Descriptor: | ALA-GLY-ALA-GLY-ALA, ALA-PHE-GLN-ALA-LYS-LEU-ASP-THR-PHE-LEU-TRP-SER, Proliferating cell nuclear antigen | | Authors: | Montoya, G, Mortuza, G.B, Blanco, F.J, Ibanez de Opakua, A. | | Deposit date: | 2015-05-14 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress.
J.Cell Biol., 212, 2016
|
|
4XOZ
 
 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, N~1~-[3-(benzyloxy)benzyl]-1H-tetrazole-1,5-diamine, SULFATE ION | | Authors: | Gelin, M, Allemand, F, Labesse, G, Guichou, J.F. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1NV5
 
 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate, Phosphate, EDTA and Thallium (5 mM) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-02 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction of Tl+ with product complexes of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
4XRL
 
 | | Crystal structure at room temperature of Erk2 in complex with an inhibitor | | Descriptor: | 1H-pyrrolo[2,3-b]pyridine-3-carbonitrile, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Gelin, M, Allemand, F, Labesse, G, Guichou, J.F. | | Deposit date: | 2015-01-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Combining 'dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7QAZ
 
 | |
4XOY
 
 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 2-amino-1,9-dihydro-6H-purine-6-thione, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Gelin, M, Allemand, F, Labesse, G, Guichou, J.F. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7ZNT
 
 | | CRYSTAL STRUCTURE OF AT7 IN COMPLEX WITH THE SECOND BROMODOMAIN OF HUMAN BRD4 AND PVHL:ELONGINC:ELONGINB | | Descriptor: | (2~{S},4~{R})-1-[(2~{R})-3-[6-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]hexylsulfanyl]-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Elongin-B, ... | | Authors: | Hughes, S.J, Casement, R, Ciulli, A. | | Deposit date: | 2022-04-22 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional E3 ligase hotspots and resistance mechanisms to small-molecule degraders.
Nat.Chem.Biol., 19, 2023
|
|
4XP0
 
 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 1H-pyrrolo[2,3-b]pyridine-3-carbonitrile, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Gelin, M, Allemand, F, Labesse, G, Guichou, J.F. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XRJ
 
 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, N~1~-[3-(benzyloxy)benzyl]-1H-tetrazole-1,5-diamine, SULFATE ION | | Authors: | Gelin, M, Allemand, F, Labesse, G, Guichou, J.F. | | Deposit date: | 2015-01-21 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8QXB
 
 | | TDP-43 amyloid fibrils: Morphology-2 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QXA
 
 | | TDP-43 amyloid fibrils: Morphology-1b | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QX9
 
 | | TDP-43 amyloid fibrils: Morphology-1a | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
5VFT
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
1R8A
 
 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide Complexes | | Descriptor: | MANGANESE (II) ION, SODIUM ION, tRNA nucleotidyltransferase | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
7AX1
 
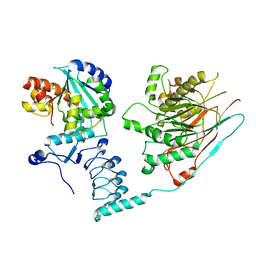 | | Crystal structure of the human CCR4-CAF1 complex | | Descriptor: | CCR4-NOT transcription complex subunit 6, CCR4-NOT transcription complex subunit 7, MAGNESIUM ION | | Authors: | Chen, Y, Khazina, E, Weichenrieder, O. | | Deposit date: | 2020-11-09 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure and functional properties of the human CCR4-CAF1 deadenylase complex.
Nucleic Acids Res., 49, 2021
|
|
8P5E
 
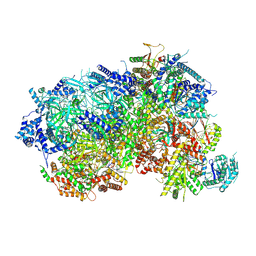 | |
8P63
 
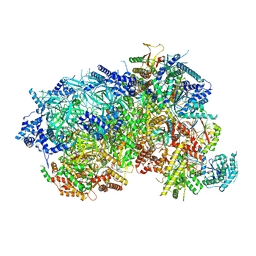 | |
8P62
 
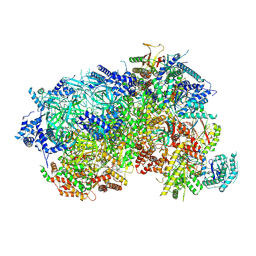 | |
1NV1
 
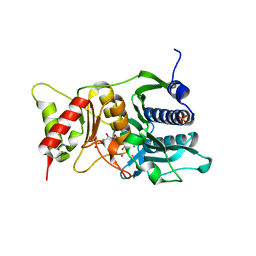 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate, Phosphate and Thallium (5 mM) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-01 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction of Tl+ with product complexes of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
5LCJ
 
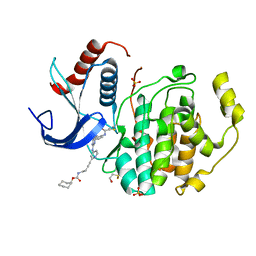 | | In-Gel Activity-Based Protein Profiling of a Clickable Covalent Erk 1/2 Inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, [(1~{R},4~{Z})-cyclooct-4-en-1-yl] ~{N}-[4-[4-[[5-chloranyl-4-[[2-(propanoylamino)phenyl]amino]pyrimidin-2-yl]amino]pyridin-2-yl]but-3-ynyl]carbamate | | Authors: | O'Reilly, M, Wright, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | In-gel activity-based protein profiling of a clickable covalent ERK1/2 inhibitor.
Mol Biosyst, 12, 2016
|
|
5LCK
 
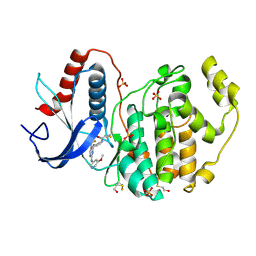 | | A Clickable Covalent ERK 1/2 Inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, ~{N}-[2-[[2-[(5-methoxypyridin-3-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl]amino]phenyl]propanamide | | Authors: | O'Reilly, M, Wright, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In-gel activity-based protein profiling of a clickable covalent ERK1/2 inhibitor.
Mol Biosyst, 12, 2016
|
|
3BRG
 
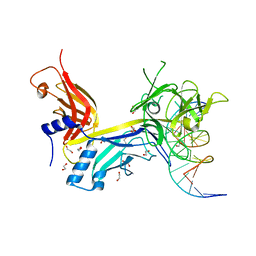 | | CSL (RBP-Jk) bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DAP*DGP*DT)-3'), DNA (5'-D(*DTP*DTP*DAP*DCP*DTP*DGP*DTP*DGP*DGP*DGP*DAP*DAP*DAP*DGP*DA)-3'), ... | | Authors: | Friedmann, D.R, Kovall, R.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
J.Biol.Chem., 283, 2008
|
|
1NV2
 
 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate, Phosphate and Thallium (20 mM) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, HYDROGENPHOSPHATE ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-01 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interaction of Tl+ with product complexes of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
1R89
 
 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide Complexes | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
4X52
 
 | | Human PARP13 (ZC3HAV1), C-Terminal PARP Domain (H810N; N830Y variant) | | Descriptor: | GLYCEROL, SULFATE ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Karlberg, T, Thorsell, A.G, Klepsch, M, Schuler, H. | | Deposit date: | 2014-12-04 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
