3TCG
 
 | | Crystal structure of E. coli OppA complexed with the tripeptide KGE | | Descriptor: | KGE Peptide, Periplasmic oligopeptide-binding protein | | Authors: | Klepsch, M.M, Kovermann, M, Low, C, Balbach, J, de Gier, J.W, Slotboom, D.J, Berntsson, R.P.-A. | | Deposit date: | 2011-08-09 | | Release date: | 2011-10-12 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Escherichia coli peptide binding protein OppA has a preference for positively charged peptides.
J.Mol.Biol., 414, 2011
|
|
3TCF
 
 | | Crystal structure of E. coli OppA complexed with endogenous ligands | | Descriptor: | Endogenous peptide, Periplasmic oligopeptide-binding protein | | Authors: | Klepsch, M.M, Kovermann, M, Low, C, Balbach, J, de Gier, J.W, Slotboom, D.J, Berntsson, R.P.-A. | | Deposit date: | 2011-08-09 | | Release date: | 2011-10-12 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Escherichia coli peptide binding protein OppA has a preference for positively charged peptides.
J.Mol.Biol., 414, 2011
|
|
3TCH
 
 | | Crystal structure of E. coli OppA in an open conformation | | Descriptor: | Periplasmic oligopeptide-binding protein | | Authors: | Klepsch, M.M, Kovermann, M, Low, C, Balbach, J, de Gier, J.W, Slotboom, D.J, Berntsson, R.P.-A. | | Deposit date: | 2011-08-09 | | Release date: | 2011-10-12 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Escherichia coli peptide binding protein OppA has a preference for positively charged peptides.
J.Mol.Biol., 414, 2011
|
|
4X52
 
 | | Human PARP13 (ZC3HAV1), C-Terminal PARP Domain (H810N; N830Y variant) | | Descriptor: | GLYCEROL, SULFATE ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Karlberg, T, Thorsell, A.G, Klepsch, M, Schuler, H. | | Deposit date: | 2014-12-04 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
2W1B
 
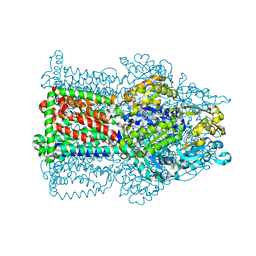 | | The structure of the efflux pump AcrB in complex with bile acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, ACRIFLAVIN RESISTANCE PROTEIN B | | Authors: | Drew, D, Klepsch, M.M, Newstead, S, Flaig, R, De Gier, J.W, Iwata, S, Beis, K. | | Deposit date: | 2008-10-17 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | The Structure of the Efflux Pump Acrb in Complex with Bile Acid.
Mol.Membr.Biol., 25, 2008
|
|
4UND
 
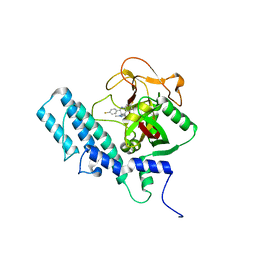 | | HUMAN ARTD1 (PARP1) - CATALYTIC DOMAIN IN COMPLEX WITH INHIBITOR TALAZOPARIB | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, POLY [ADP-RIBOSE] POLYMERASE 1, SODIUM ION | | Authors: | Karlberg, T, Thorsell, A.G, Ekblad, T, Klepsch, M, Pinto, A.F, Tresaugues, L, Moche, M, Schuler, H. | | Deposit date: | 2014-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
