1WYW
 
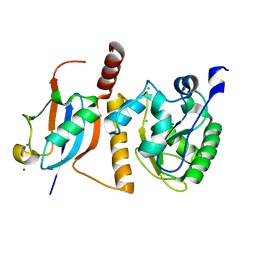 | | Crystal Structure of SUMO1-conjugated thymine DNA glycosylase | | Descriptor: | CHLORIDE ION, G/T mismatch-specific thymine DNA glycosylase, MAGNESIUM ION, ... | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-02-17 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymine DNA glycosylase conjugated to SUMO-1.
Nature, 435, 2005
|
|
6OEQ
 
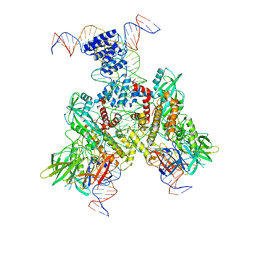 | | Cryo-EM structure of mouse RAG1/2 12RSS-PRC/23RSS-NFC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEP
 
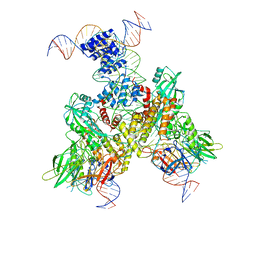 | | Cryo-EM structure of mouse RAG1/2 12RSS-NFC/23RSS-PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEN
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OER
 
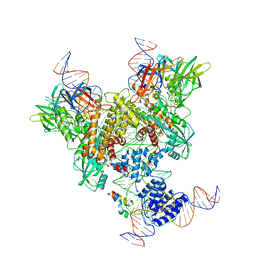 | | Cryo-EM structure of mouse RAG1/2 NFC complex (DNA2) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEO
 
 | | Cryo-EM structure of mouse RAG1/2 NFC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2W3O
 
 | |
8JYD
 
 | |
2D07
 
 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | Descriptor: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-07-26 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
6T7F
 
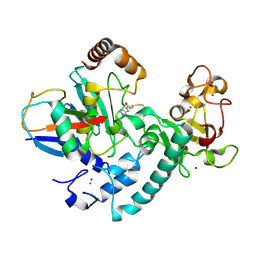 | | RCR E3 ligase E2-Ubiquitin transthiolation intermediate | | Descriptor: | 3,3-bis(sulfanyl)-~{N}-(1~{H}-1,2,3-triazol-4-ylmethyl)propanamide, E3 ubiquitin-protein ligase MYCBP2, Polyubiquitin-C, ... | | Authors: | Mabbitt, P.D, Virdee, S. | | Deposit date: | 2019-10-21 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis for RING-Cys-Relay E3 ligase activity and its role in axon integrity.
Nat.Chem.Biol., 16, 2020
|
|
6DBR
 
 | | Cryo-EM structure of RAG in complex with one melted RSS and one unmelted RSS | | Descriptor: | CALCIUM ION, Forward strand of melted RSS substrate DNA, Forward strand of unmelted RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBI
 
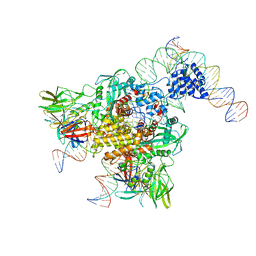 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS signal end, Forward strand of 23-RSS signal end, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBJ
 
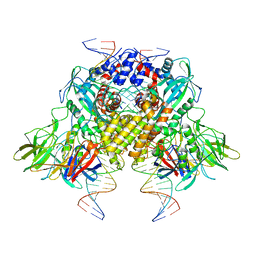 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward stand of RSS signal end, Forward strand of coding flank, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6V0V
 
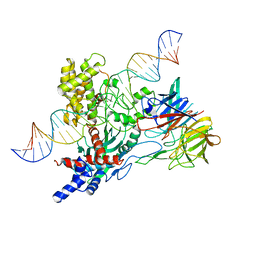 | | Cryo-EM structure of mouse WT RAG1/2 NFC complex (DNA0) | | Descriptor: | CALCIUM ION, DNA (30-MER), V(D)J recombination-activating protein 1, ... | | Authors: | Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2019-11-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7TJF
 
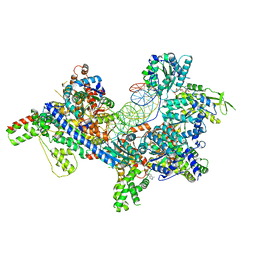 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA, 84 bp ARS1, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
8DMB
 
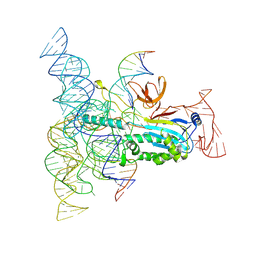 | | Structure of Desulfovirgula thermocuniculi IsrB (DtIsrB) in complex with omega RNA and target DNA | | Descriptor: | MAGNESIUM ION, Ubiquitin-like protein SMT3,IsrB protein,monomeric superfolder Green Fluorescent Protein, non-target DNA, ... | | Authors: | Seiichi, H, Kappel, K, Zhang, F. | | Deposit date: | 2022-07-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the OMEGA nickase IsrB in complex with omega RNA and target DNA.
Nature, 610, 2022
|
|
5G4Q
 
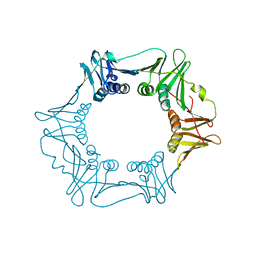 | | H.pylori Beta clamp in complex with 5-chloroisatin | | Descriptor: | 5-chloro-1H-indole-2,3-dione, DNA POLYMERASE III SUBUNIT BETA | | Authors: | Pandey, P, Gourinath, S. | | Deposit date: | 2016-05-16 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening of E. coli beta-clamp Inhibitors Revealed that Few Inhibit Helicobacter pylori More Effectively: Structural and Functional Characterization.
Antibiotics (Basel), 7, 2018
|
|
5G48
 
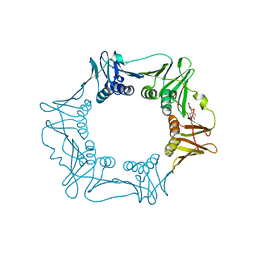 | | H.pylori Beta clamp in complex with Diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, DNA POLYMERASE III SUBUNIT BETA | | Authors: | Pandey, P, Gourinath, S. | | Deposit date: | 2016-05-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Targeting the beta-clamp in Helicobacter pylori with FDA-approved drugs reveals micromolar inhibition by diflunisal.
FEBS Lett., 591, 2017
|
|
6KHY
 
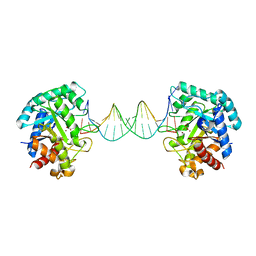 | | The crystal structure of AsfvAP:AG | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (AGCGTCACCGACGAGGC), DNA(AGCGTCACCGACGAGG), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2019-07-16 | | Release date: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | A unique DNA-binding mode of African swine fever virus AP endonuclease.
Cell Discov, 6, 2020
|
|
2QHO
 
 | |
3V8K
 
 | |
3V7R
 
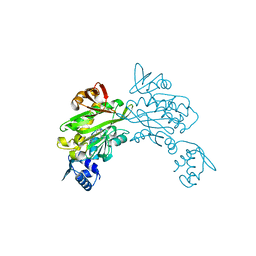 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | (3aS,4S,6aR)-4-(5-{1-[4-(6-amino-9H-purin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}pentyl)tetrahydro-1H-thieno[3,4-d]imidazol-2(3H)-one, Biotin ligase | | Authors: | Yap, M.Y, Pendini, N.R. | | Deposit date: | 2011-12-21 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
3V8L
 
 | |
3V7S
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor 0364 | | Descriptor: | 5-methyl-3-[4-(4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)butyl]-1,3-benzoxazol-2(3H)-one, Biotin ligase | | Authors: | Yap, M.Y, Pendini, N.R. | | Deposit date: | 2011-12-21 | | Release date: | 2012-04-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
4DQ2
 
 | | Structure of staphylococcus aureus biotin protein ligase in complex with biotinol-5'-amp | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, Biotin-[acetyl-CoA-carboxylase] ligase | | Authors: | Wilce, M, Yap, M, Pendini, N, Soares de Costa, T, Polyak, S, Tieu, W, Booker, G, Wallace, J. | | Deposit date: | 2012-02-14 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
