3DWV
 
 | |
2N70
 
 | | Two-fold symmetric structure of the 18-60 construct of S31N M2 from Influenza A in lipid bilayers | | Descriptor: | Matrix protein 2 | | Authors: | Andreas, L.B, Reese, M, Eddy, M.T, Gelev, V, Ni, Q, Miller, E.A, Emsley, L, Pintacuda, G, Chou, J.J, Griffin, R.G. | | Deposit date: | 2015-09-01 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure and Mechanism of the Influenza A M218-60 Dimer of Dimers.
J.Am.Chem.Soc., 137, 2015
|
|
2N1M
 
 | |
2NVD
 
 | | Human Aldose Reductase complexed with novel naphtho[1,2-d]isothiazole acetic acid derivative (2) | | Descriptor: | 2-(CARBOXYMETHYL)-1-OXO-1,2-DIHYDRONAPHTHO[1,2-D]ISOTHIAZOLE-4-CARBOXYLIC ACID 3,3-DIOXIDE, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2006-11-12 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Evidence for a novel binding site conformer of aldose reductase in ligand-bound state
J.Mol.Biol., 369, 2007
|
|
2NVC
 
 | | Human Aldose Reductase complexed with novel naphtho[1,2-d]isothiazole acetic acid derivative (3) | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {4-[(CARBOXYMETHOXY)CARBONYL]-3,3-DIOXIDO-1-OXONAPHTHO[1,2-D]ISOTHIAZOL-2(1H)-YL}ACETIC ACID | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2006-11-12 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Evidence for a novel binding site conformer of aldose reductase in ligand-bound state
J.Mol.Biol., 369, 2007
|
|
1M0M
 
 | | BACTERIORHODOPSIN M1 INTERMEDIATE AT 1.43 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN, ... | | Authors: | Lanyi, J.K. | | Deposit date: | 2002-06-13 | | Release date: | 2002-09-11 | | Last modified: | 2015-09-09 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystallographic structure of the retinal and the protein after deprotonation of the Schiff base: the switch in the bacteriorhodopsin photocycle.
J.Mol.Biol., 321, 2002
|
|
1MAC
 
 | | CRYSTAL STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF BACILLUS MACERANS ENDO-1,3-1,4-BETA-GLUCANASE | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-beta-glucanase.
J.Biol.Chem., 270, 1995
|
|
6LF9
 
 | | Crystal structure of pSLA-1*1301 complex with dodecapeptide RVEDVTNTAEYW | | Descriptor: | ARG-VAL-GLU-ASP-VAL-THR-ASN-THR-ALA-GLU-TYR-TRP, Beta-2-microglobulin, MHC class I antigen | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-11-30 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptidomes and Structures Illustrate How SLA-I Micropolymorphism Influences the Preference of Binding Peptide Length.
Front Immunol, 2022
|
|
6LF8
 
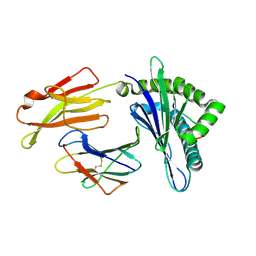 | | Crystal structure of pSLA-1*0401 complex with dodecapeptide RVEDVTNTAEYW | | Descriptor: | ARG-VAL-GLU-ASP-VAL-THR-ASN-THR-ALA-GLU-TYR-TRP, Beta-2-microglobulin, MHC class I antigen | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-11-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Peptidomes of Swine MHC Class I with Long Peptides Reveal the Cross-Species Characteristics of the Novel N-Terminal Extension Presentation Mode.
J Immunol., 208, 2022
|
|
1KWG
 
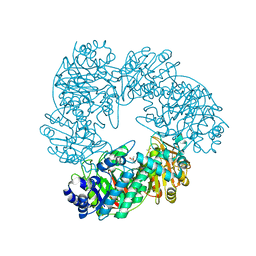 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trimeric Crystal Structure of the Glycoside Hydrolase Family 42 beta-Galactosidase from Thermus thermophilus A4 and the Structure of its Complex with Galactose
J.MOL.BIOL., 322, 2002
|
|
1KWK
 
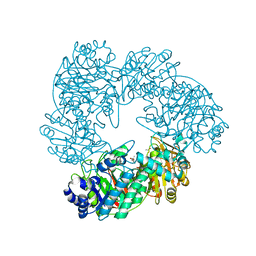 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase in complex with galactose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trimeric crystal structure of the glycoside hydrolase family 42 beta-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose.
J.Mol.Biol., 322, 2002
|
|
8FHD
 
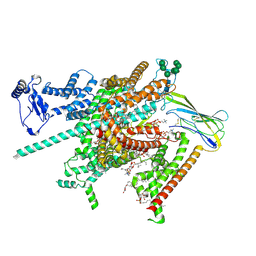 | | Cryo-EM structure of human voltage-gated sodium channel Nav1.6 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (5E,17R,20S)-23-amino-20-hydroxy-14,20-dioxo-15,19,21-trioxa-20lambda~5~-phosphatricos-5-en-17-yl hexadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of human voltage-gated sodium channel Na v 1.6.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3A7Q
 
 | | Structural basis for specific recognition of reelin by its receptors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yasui, N, Nogi, T, Takagi, J. | | Deposit date: | 2009-10-01 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Specific Recognition of Reelin by Its Receptors
Structure, 18, 2010
|
|
1U1H
 
 | | A. thaliana cobalamine independent methionine synthase | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, METHIONINE, SULFATE ION, ... | | Authors: | Ferrer, J.-L, Ravanel, S, Robert, M, Dumas, R. | | Deposit date: | 2004-07-15 | | Release date: | 2004-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of cobalamin-independent methionine synthase complexed with zinc, homocysteine, and methyltetrahydrofolate
J.Biol.Chem., 279, 2004
|
|
1U22
 
 | | A. thaliana cobalamine independent methionine synthase | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, ... | | Authors: | Ferrer, J.-L, Ravanel, S, Robert, M, Dumas, R. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of cobalamin-independent methionine synthase complexed with zinc, homocysteine, and methyltetrahydrofolate
J.Biol.Chem., 279, 2004
|
|
1WP6
 
 | | Crystal structure of maltohexaose-producing amylase from alkalophilic Bacillus sp.707. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Glucan 1,4-alpha-maltohexaosidase, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2004-08-31 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and crystallographic analyses of maltohexaose-producing amylase from alkalophilic Bacillus sp. 707
Biochemistry, 43, 2004
|
|
1W8S
 
 | | The mechanism of the Schiff Base Forming Fructose-1,6-bisphosphate Aldolase: Structural analysis of reaction intermediates | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2004-09-27 | | Release date: | 2005-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of the Schiff base forming fructose-1,6-bisphosphate aldolase: structural analysis of reaction intermediates.
Biochemistry, 44, 2005
|
|
8GNG
 
 | | Crystal structure of human adenosine A2A receptor in complex with istradefylline. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 8-[(~{E})-2-(3,4-dimethoxyphenyl)ethenyl]-1,3-diethyl-7-methyl-purine-2,6-dione, Adenosine receptor A2a, ... | | Authors: | Suzuki, M, Saito, J, Miyagi, H, Yasunaga, M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | In Vitro Pharmacological Profile of KW-6356, a Novel Adenosine A 2A Receptor Antagonist/Inverse Agonist.
Mol.Pharmacol., 103, 2023
|
|
8GNE
 
 | | Crystal structure of human adenosine A2A receptor in complex with an insurmountable inverse agonist, KW-6356. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Suzuki, M, Saito, J, Miyagi, H, Yasunaga, M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In Vitro Pharmacological Profile of KW-6356, a Novel Adenosine A 2A Receptor Antagonist/Inverse Agonist.
Mol.Pharmacol., 103, 2023
|
|
1VJS
 
 | |
1WPC
 
 | | Crystal structure of maltohexaose-producing amylase complexed with pseudo-maltononaose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2004-09-01 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and crystallographic analyses of maltohexaose-producing amylase from alkalophilic Bacillus sp. 707
Biochemistry, 43, 2004
|
|
1XF0
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 (AKR1C3) complexed with delta4-androstene-3,17-dione and NADP | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, ACETATE ION, Aldo-keto reductase family 1 member C3, ... | | Authors: | Qiu, W, Zhou, M, Labrie, F, Lin, S.-X. | | Deposit date: | 2004-09-13 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the multispecific 17beta-hydroxysteroid dehydrogenase type 5: critical androgen regulation in human peripheral tissues
Mol.Endocrinol., 18, 2004
|
|
1XGD
 
 | | Apo R268A human aldose reductase | | Descriptor: | Aldose reductase | | Authors: | Brownlee, J.M, Bohren, K.M, Milne, A.C, Gabbay, K.H, Harrison, D.H.T. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of Apo R268A human aldose reductase: Hinges and latches that control the kinetic mechanism
Biochim.Biophys.Acta, 1748, 2005
|
|
1T7C
 
 | | CRYSTAL STRUCTURE OF THE P1 GLU BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Czapinska, H, Helland, R, Otlewski, J, Smalas, A.O. | | Deposit date: | 2004-05-09 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of five bovine chymotrypsin complexes with P1 BPTI variants.
J.Mol.Biol., 344, 2004
|
|
1T8M
 
 | | CRYSTAL STRUCTURE OF THE P1 HIS BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Czapinska, H, Helland, R, Otlewski, J, Smalas, A.O. | | Deposit date: | 2004-05-13 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of five bovine chymotrypsin complexes with P1 BPTI variants.
J.Mol.Biol., 344, 2004
|
|
