3U0T
 
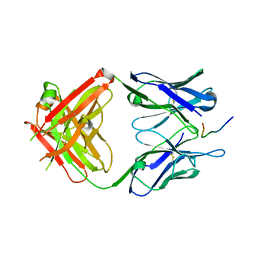 | | Fab-antibody complex | | Descriptor: | Amyloid beta A4 protein, ponezumab HC Fab, ponezumab LC Fab | | Authors: | LaPorte, S.L, Pons, J.P. | | Deposit date: | 2011-09-29 | | Release date: | 2012-01-11 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of C-terminal beta-Amyloid Peptide Binding by the Antibody Ponezumab for the Treatment of Alzheimer's Disease
J.Mol.Biol., 421, 2012
|
|
8SEK
 
 | |
8SEL
 
 | |
6SZF
 
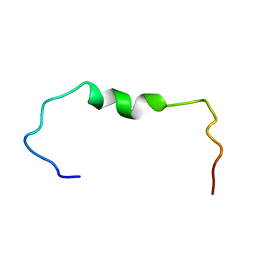 | | Solution structure of the amyloid beta-peptide (1-42) | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Grimaldi, M, Santoro, A, Stillitano, I, Buonocore, M, D'Ursi, A.M. | | Deposit date: | 2019-10-02 | | Release date: | 2020-10-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Exploring the Early Stages of the Amyloid A beta (1-42) Peptide Aggregation Process: An NMR Study.
Pharmaceuticals, 14, 2021
|
|
6TI7
 
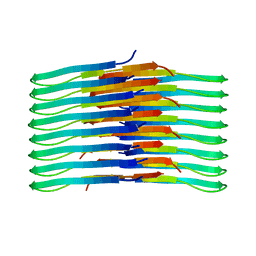 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
3UMH
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with cadmium | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
3SV1
 
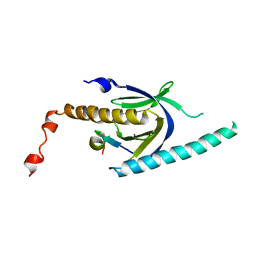 | | Crystal structure of APP peptide bound rat Mint2 PARM | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2, Amyloid beta A4 protein | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
6TI5
 
 | | A New Structural Model of Abeta(1-40) Fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Bertini, I, Gonnelli, L, Luchinat, C, Mao, J, Nesi, A. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6TI6
 
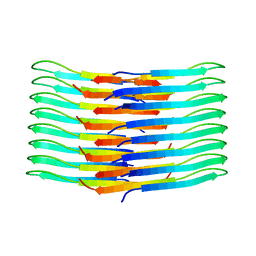 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
1Z0Q
 
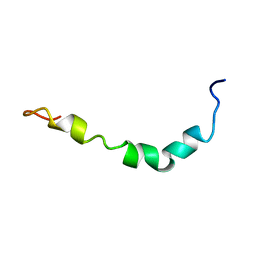 | | Aqueous Solution Structure of the Alzheimer's Disease Abeta Peptide (1-42) | | Descriptor: | Alzheimer's disease amyloid | | Authors: | Tomaselli, S, Esposito, V, Vangone, P, van Nuland, N.A, Bonvin, A.M, Guerrini, R, Tancredi, T, Temussi, P.A, Picone, D. | | Deposit date: | 2005-03-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The alpha-to-beta Conformational Transition of Alzheimer's Abeta-(1-42) Peptide in Aqueous Media is Reversible: A Step by Step Conformational Analysis Suggests the Location of beta Conformation Seeding
Chembiochem, 7, 2006
|
|
3UMK
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with copper | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION, ... | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
1ZE9
 
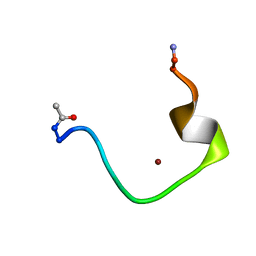 | | Zinc-binding domain of Alzheimer's disease amyloid beta-peptide complexed with a zinc (II) cation | | Descriptor: | 16-mer from Alzheimer's disease amyloid Protein, ZINC ION | | Authors: | Zirah, S, Kozin, S.A, Mazur, A.K, Blond, A, Cheminant, M, Segalas-Milazzo, I, Debey, P, Rebuffat, S. | | Deposit date: | 2005-04-18 | | Release date: | 2005-05-03 | | Last modified: | 2013-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural changes of region 1-16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging.
J.Biol.Chem., 281, 2006
|
|
1ZJD
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with Kunitz Protease Inhibitor Domain of Protease Nexin II | | Descriptor: | Catalytic Domain of Coagulation Factor XI, Kunitz Protease Inhibitory Domain of Protease Nexin II | | Authors: | Jin, L, Navaneetham, D, Pandey, P, Strickler, J.E, Babine, R.E, Walsh, P.N, Abdel-Meguid, S.S. | | Deposit date: | 2005-04-28 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mutational Analyses of the Molecular Interactions between the Catalytic Domain of Factor XIa and the Kunitz Protease Inhibitor Domain of Protease Nexin 2
J.Biol.Chem., 280, 2005
|
|
3UMI
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with zinc | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION, ... | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
1ZE7
 
 | | Zinc-binding domain of Alzheimer's disease amyloid beta-peptide in water solution at pH 6.5 | | Descriptor: | 16-mer from Alzheimer's disease amyloid Protein | | Authors: | Zirah, S, Kozin, S.A, Mazur, A.K, Blond, A, Cheminant, M, Segalas-Milazzo, I, Debey, P, Rebuffat, S. | | Deposit date: | 2005-04-18 | | Release date: | 2005-05-03 | | Last modified: | 2013-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural changes of region 1-16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging.
J.Biol.Chem., 281, 2006
|
|
3KTM
 
 | | Structure of the Heparin-induced E1-Dimer of the Amyloid Precursor Protein (APP) | | Descriptor: | (3R)-butane-1,3-diol, ACETATE ION, Amyloid beta A4 protein, ... | | Authors: | Dahms, S.O, Hoefgen, S, Roeser, D, Schlott, B, Guhrs, K.H, Than, M.E. | | Deposit date: | 2009-11-25 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and biochemical analysis of the heparin-induced E1 dimer of the amyloid precursor protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3L81
 
 | | Crystal structure of adaptor protein complex 4 (AP-4) mu4 subunit C-terminal domain, in complex with a sorting peptide from the amyloid precursor protein (APP) | | Descriptor: | AP-4 complex subunit mu-1, Amyloid beta A4 protein, GLYCEROL | | Authors: | Mardones, G.A, Rojas, A.L, Burgos, P.V, Dasilva, L.L.P, Prabhu, Y, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2009-12-29 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sorting of the Alzheimer's disease amyloid precursor protein mediated by the AP-4 complex.
Dev.Cell, 18, 2010
|
|
3MXC
 
 | |
3MXY
 
 | |
3NYL
 
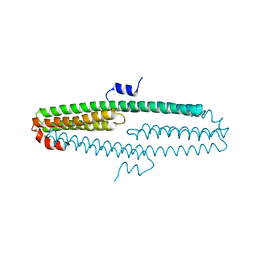 | | The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain | | Descriptor: | Amyloid beta (A4) protein (Peptidase nexin-II, Alzheimer disease), isoform CRA_b | | Authors: | Ha, Y, Hu, J, Lee, S, Liu, X, Wang, Y. | | Deposit date: | 2010-07-15 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain.
Mol.Cell, 15, 2004
|
|
3L33
 
 | | Human mesotrypsin complexed with amyloid precursor protein inhibitor(APPI) | | Descriptor: | Amyloid beta A4 protein, CALCIUM ION, FORMIC ACID, ... | | Authors: | Salameh, M.A, Soares, A.S, Radisky, E.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Determinants of affinity and proteolytic stability in interactions of Kunitz family protease inhibitors with mesotrypsin.
J.Biol.Chem., 285, 2010
|
|
3OVJ
 
 | |
3PZZ
 
 | |
3OW9
 
 | |
3Q2X
 
 | |
