7YU4
 
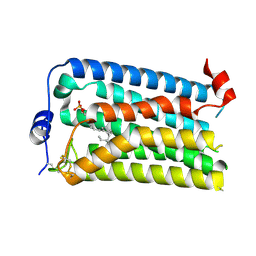 | |
7YU7
 
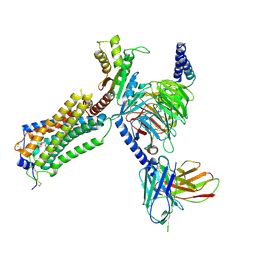 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state3 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
5JZL
 
 | | The Structure of Monomeric Ultra Stable Green Fluorescent Protein | | Descriptor: | CHLORIDE ION, Green fluorescent protein, SODIUM ION | | Authors: | Gunn, N.J, Yong, K.J, Scott, D.J, Griffin, M.D.W. | | Deposit date: | 2016-05-17 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel Ultra-Stable, Monomeric Green Fluorescent Protein For Direct Volumetric Imaging of Whole Organs Using CLARITY.
Sci Rep, 8, 2018
|
|
7Z2C
 
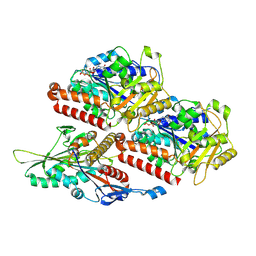 | | P. falciparum kinesin-8B motor domain in no nucleotide bound to tubulin dimer | | Descriptor: | Detyrosinated tubulin alpha-1B chain, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein, ... | | Authors: | Liu, T, Shilliday, F, Cook, A.D, Moores, C.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Nat Commun, 13, 2022
|
|
4ZG0
 
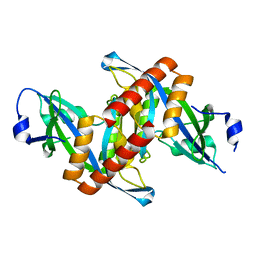 | | Crystal structure of Mouse Syndesmos protein | | Descriptor: | Protein syndesmos | | Authors: | Lee, I, Kim, H, Yoo, J, Cho, H, Lee, W. | | Deposit date: | 2015-04-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of syndesmos and its interaction with Syndecan-4 proteoglycan
Biochem.Biophys.Res.Commun., 463, 2015
|
|
7UV5
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S/D286N mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Papain-like protease nsp3, Ubiquitin, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-29 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
6XHB
 
 | | Crystal Structure of wild-type KRAS (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF (crystal form II) | | Descriptor: | CHLORIDE ION, GLYCEROL, GTPase KRas, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
6XR3
 
 | |
6XRR
 
 | | Structure of SciW bound to the Rhs1 Transmembrane Domain from Salmonella typhimurium | | Descriptor: | Putative cytoplasmic protein, RHS repeat protein, SULFATE ION | | Authors: | Sachar, K, Ahmad, S, Whitney, J.C, Prehna, G. | | Deposit date: | 2020-07-13 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for effector transmembrane domain recognition by type VI secretion system chaperones.
Elife, 9, 2020
|
|
6WVT
 
 | | Structural basis of alphaE-catenin - F-actin catch bond behavior | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Xu, X.P, Pokutta, S, Torres, M, Swift, M.F, Hanein, D, Volkmann, N, Weis, W.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of alpha E-catenin-F-actin catch bond behavior.
Elife, 9, 2020
|
|
7D6F
 
 | | The crystal structure of ARMS-PBM/MAGI2-PDZ4 | | Descriptor: | GLYCEROL, Kinase D-interacting substrate of 220 kDa, Membrane-associated guanylate kinase, ... | | Authors: | Ye, J, Zhang, Y, Zhong, Z, Wang, C. | | Deposit date: | 2020-09-30 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal structure of the PDZ4 domain of MAGI2 in complex with PBM of ARMS reveals a canonical PDZ recognition mode.
Neurochem.Int., 149, 2021
|
|
5IY5
 
 | | Electron transfer complex of cytochrome c and cytochrome c oxidase at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, S, Baba, J, Aoe, S, Shimada, A, Yamashita, E, Tsukihara, T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode
EMBO J., 36, 2017
|
|
8AT5
 
 | | native Coxsackievirus A9 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Domanska, A, Plavec, Z, Ruokolainen, V, Marjomaki, V.S, Butcher, S.J. | | Deposit date: | 2022-08-22 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Studies Reveal that Endosomal Cations Promote Formation of Infectious Coxsackievirus A9 A-Particles, Facilitating RNA and VP4 Release.
J.Virol., 96, 2022
|
|
5U1J
 
 | | Structure of pNOB8 ParA bound to nonspecific DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*TP*AP*AP*TP*GP*AP*CP*GP*CP*CP*GP*GP*CP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*GP*CP*CP*GP*GP*CP*GP*TP*CP*AP*TP*GP*AP*CP*AP*CP*G)-3'), PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-04-19 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of partition protein ParA with nonspecific DNA and ParB effector reveal molecular insights into principles governing Walker-box DNA segregation.
Genes Dev., 31, 2017
|
|
6X91
 
 | | Crystal structure of MBP-fused human APOBEC1 | | Descriptor: | CACODYLATE ION, Maltodextrin-binding protein, C->U-editing enzyme APOBEC-1 chimera, ... | | Authors: | Wolfe, A.D, Li, S.-X, Chen, X.S. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The structure of APOBEC1 and insights into its RNA and DNA substrate selectivity.
NAR Cancer, 2, 2020
|
|
4ZD3
 
 | | Structure of a transglutaminase 2-specific autoantibody Fab fragment | | Descriptor: | 679-14-14E06 Fab fragment heavy chain, 679-14-14E06 Fab fragment light chain | | Authors: | Chen, X, Dalhus, B, Hnida, K, Iversen, R, Sollid, L.M. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Antigen Recognition by Transglutaminase 2-specific Autoantibodies in Celiac Disease.
J.Biol.Chem., 290, 2015
|
|
8ADQ
 
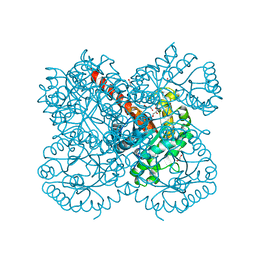 | | Crystal structure of holo-SwHPA-Mg (hydroxy ketone aldolase) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate and D-Glyceraldehyde | | Descriptor: | 3-HYDROXYPYRUVIC ACID, BROMIDE ION, D-Glyceraldehyde, ... | | Authors: | Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5JMR
 
 | | X-ray structure of the furin inhibitory antibody Nb14 | | Descriptor: | CHLORIDE ION, camelid VHH fragment | | Authors: | Dahms, S.O, Than, M.E. | | Deposit date: | 2016-04-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The structure of a furin-antibody complex explains non-competitive inhibition by steric exclusion of substrate conformers.
Sci Rep, 6, 2016
|
|
1UJ3
 
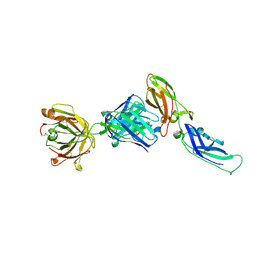 | | Crystal structure of a humanized Fab fragment of anti-tissue-factor antibody in complex with tissue factor | | Descriptor: | IgG Fab heavy chain, IgG Fab light chain, tissue factor | | Authors: | Ohto, U, Mizutani, R, Nakamura, M, Adachi, H, Satow, Y. | | Deposit date: | 2003-07-25 | | Release date: | 2004-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a humanized Fab fragment of anti-tissue-factor antibody in complex with tissue factor.
J.Synchrotron Radiat., 11, 2004
|
|
6XKW
 
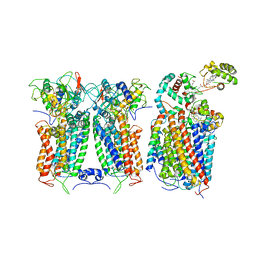 | | R. capsulatus CIII2CIV bipartite super-complex (SC-2A) with CcoH/cy | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
5U4W
 
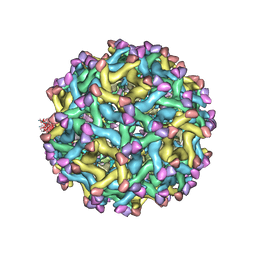 | | Cryo-EM Structure of Immature Zika Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein, ... | | Authors: | Mangala Prasad, V, Miller, A.S, Klose, T, Sirohi, D, Buda, G, Jiang, W, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Structure of the immature Zika virus at 9 angstrom resolution.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5U5N
 
 | |
7DC7
 
 | | Crystal structure of D12 Fab-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D12 Fab heavy chain, D12 Fab light chain | | Authors: | Kawauchi, H, Fukami, T.A, Tatsumi, K, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
7L9P
 
 | | Structure of human SHLD2-SHLD3-REV7-TRIP13(E253Q) complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Pachytene checkpoint protein 2 homolog, ... | | Authors: | Xie, W, Patel, D.J. | | Deposit date: | 2021-01-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5TQ1
 
 | |
