6N2W
 
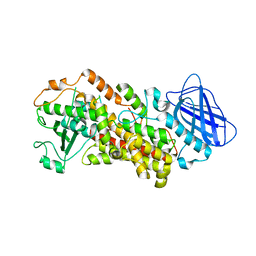 | | The structure of Stable-5-Lipoxygenase bound to NDGA | | Descriptor: | 4-[(2R,3S)-3-[(3,4-DIHYDROXYPHENYL)METHYL]-2-METHYLBUTYL]BENZENE-1,2-DIOL, Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Newcomer, M.E, Gilbert, N.C, Neau, D.B. | | Deposit date: | 2018-11-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products.
Nat.Chem.Biol., 16, 2020
|
|
6G0E
 
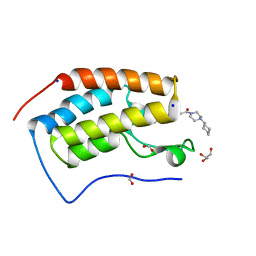 | | BRD4 (BD1) in complex with APSC-derived ligands | | Descriptor: | 4-[2-(4-cyclohexylpiperazin-4-ium-1-yl)-2-oxidanylidene-ethyl]sulfanyl-1-ethyl-quinolin-2-one, Bromodomain-containing protein 4, FORMIC ACID, ... | | Authors: | Pretzel, J, Humbeck, L. | | Deposit date: | 2018-03-18 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.613 Å) | | Cite: | Discovery of an Unexpected Similarity in Ligand Binding Between BRD4 and PPARgamma
Chemrxiv, 2019
|
|
6XTL
 
 | |
6PZN
 
 | |
5EBB
 
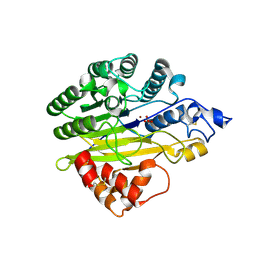 | | Structure of human sphingomyelinase phosphodiesterase like 3A (SMPDL3A) with Zn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, GLYCEROL, ... | | Authors: | Lim, S.M, Yeung, K, Tresaugues, L, Teo, H.L, Nordlund, P. | | Deposit date: | 2015-10-19 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a - an acid sphingomyelinase homologue with a novel nucleotide hydrolase activity.
Febs J., 283, 2016
|
|
6XTR
 
 | |
7D2C
 
 | | The Crystal Structure of human PARP14 from Biortus. | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP14 | | Authors: | Wang, F, Miao, Q, Lv, Z, Cheng, W, Lin, D, Xu, X, Tan, J. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The Crystal Structure of human PARP14 from Biortus.
To Be Published
|
|
5L47
 
 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with cyanoselenobarbital (seleniated barbiturate) | | Descriptor: | 2-[5-ethyl-2,4,6-tris(oxidanylidene)-1,3-diazinan-5-yl]ethyl selenocyanate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Reinholds Ruza, R, Fourati, Z, Delarue, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Barbiturates Bind in the GLIC Ion Channel Pore and Cause Inhibition by Stabilizing a Closed State.
J. Biol. Chem., 292, 2017
|
|
7D42
 
 | |
8P61
 
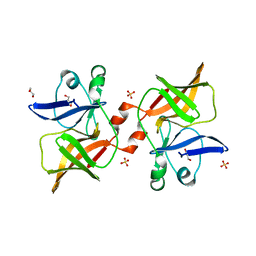 | | Crystal structure of O'nyong'nyong virus capsid protease (106-256) | | Descriptor: | Capsid protein, GLYCEROL, SULFATE ION | | Authors: | Plewka, J, Chykunova, Y, Wilk, P, Sienczyk, M, Dubin, G, Pyrc, K. | | Deposit date: | 2023-05-24 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autoinhibition of suicidal capsid protease from O'nyong'nyong virus.
Int.J.Biol.Macromol., 262, 2024
|
|
5W46
 
 | |
6N4G
 
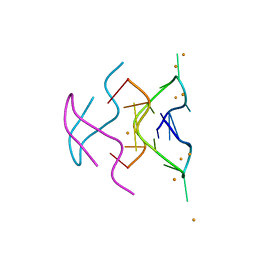 | |
8OZ4
 
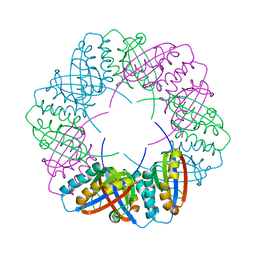 | | Populus tremula stable protein 1 with an alternate crystal lattice | | Descriptor: | Stable protein 1 | | Authors: | Sklyar, J, Zeibaq, Y, Bachar, O, Yehezkeli, O, Adir, N. | | Deposit date: | 2023-05-08 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Bioengineered Stable Protein 1-Hemin Complex with Enhanced Peroxidase-Like Catalytic Properties
Small Science, 2024
|
|
5VVJ
 
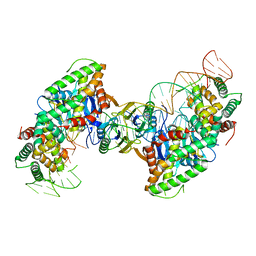 | | Cas1-Cas2 bound to half-site intermediate | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (112-MER), ... | | Authors: | Wright, A.V, Knott, G.J, Doxzen, K.W, Doudna, J.A. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
6XV4
 
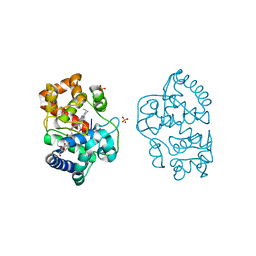 | | Neutron structure of ferric ascorbate peroxidase-ascorbate complex | | Descriptor: | ASCORBIC ACID, Ascorbate peroxidase, POTASSIUM ION, ... | | Authors: | Kwon, H, Basran, J, Devos, J.M, Schrader, T.E, Ostermann, A, Blakeley, M.P, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2020-01-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Visualizing the protons in a metalloenzyme electron proton transfer pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XVB
 
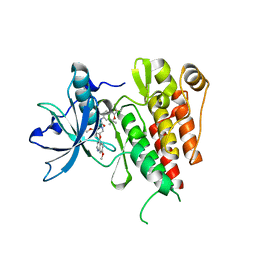 | | Crystal structure of the kinase domain of human c-KIT with a cyclic imidate inhibitor covalently bound to Cys788 | | Descriptor: | Mast/stem cell growth factor receptor Kit,Mast/stem cell growth factor receptor Kit, ~{N}-(4,4-dimethyl-2-propyl-3,1-benzoxazin-6-yl)-2-[3-methoxy-5-(7-methoxyquinolin-4-yl)oxy-pyridin-2-yl]ethanamide | | Authors: | Schimpl, M, McAuley, K, Hoyt, E.A, Thomas, M, Bodnarchuk, M.S, Lewis, H.J, Barratt, D, Deery, M.J, Ogg, D.J, Bernardes, G.J.L, Ward, R.A, Kettle, J.G, Waring, M.J. | | Deposit date: | 2020-01-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Alkynyl Benzoxazines and Dihydroquinazolines as Cysteine Targeting Covalent Warheads and Their Application in Identification of Selective Irreversible Kinase Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
7D6H
 
 | |
5HWQ
 
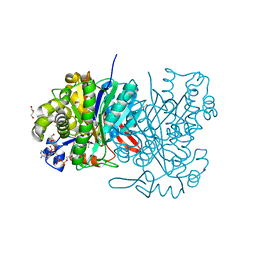 | | MvaS in complex with acetoacetyl coenzyme A | | Descriptor: | ACETOACETYL-COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
6MY4
 
 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98M,HC-G99M,LC-S30bR] | | Descriptor: | 1,2-ETHANEDIOL, anti-VEGF-A Fab fragment bH1 heavy chain, anti-VEGF-A Fab fragment bH1 light chain | | Authors: | Shi, R, Picard, M.-E, Manenda, M. | | Deposit date: | 2018-11-01 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
6XVF
 
 | | Crystal structure of bovine cytochrome bc1 in complex with tetrahydro-quinolone inhibitor JAG021 | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Amporndanai, K, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent Tetrahydroquinolone Eliminates Apicomplexan Parasites.
Front Cell Infect Microbiol, 10, 2020
|
|
6PUF
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-5'D-5-OP-RU | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
5DUQ
 
 | | Active human c1-inhibitor in complex with dextran sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasma protease C1 inhibitor, SULFITE ION, ... | | Authors: | Dijk, M, Holkers, J, Voskamp, P, Giannetti, B.M, Waterreus, W.J, van Veen, H.A, Pannu, N.S. | | Deposit date: | 2015-09-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | How Dextran Sulfate Affects C1-inhibitor Activity: A Model for Polysaccharide Potentiation.
Structure, 24, 2016
|
|
6G16
 
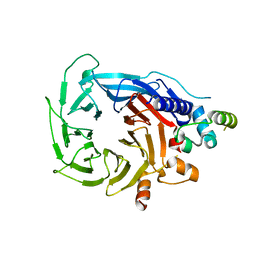 | | Structure of the human RBBP4:MTA1(464-546) complex showing loop exchange | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
5KM6
 
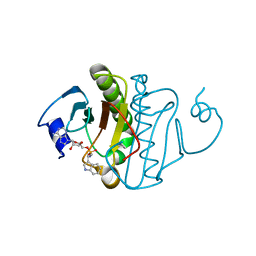 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant Ara-A nucleoside phosphoramidate substrate complex | | Descriptor: | Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[2-(1~{H}-indol-3-yl)ethyl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
8P37
 
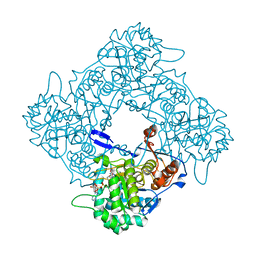 | | Structure a catalytically inactive mutant of the IMP dehydrogenase related protein GUAB3 from Synechocystis PCC 6803 | | Descriptor: | IMP dehydrogenase subunit, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Hernandez-Gomez, A, Fernandez-Justel, D, Buey, R.M. | | Deposit date: | 2023-05-17 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | GuaB3, an overlooked enzyme in cyanobacteria's toolbox that sheds light on IMP dehydrogenase evolution.
Structure, 31, 2023
|
|
