8Q6M
 
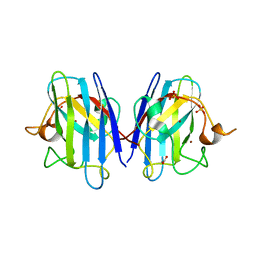 | | Human SOD1 low dose data collecton | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Hossain, A, Agar, J.N, Hasnain, S.S. | | Deposit date: | 2023-08-14 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Evaluating protein cross-linking as a therapeutic strategy to stabilize SOD1 variants in a mouse model of familial ALS.
Plos Biol., 22, 2024
|
|
8B61
 
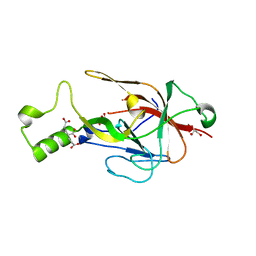 | | Crystal structure of BfrC protein from Bacteroides fragilis NCTC 9343 | | Descriptor: | Conserved hypothetical lipoprotein, GLYCEROL, pentane-1,3,5-tricarboxylic acid | | Authors: | Antonyuk, S.V, Barnett, K, Strange, R.W, Olczak, T. | | Deposit date: | 2022-09-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Bacteroides fragilis expresses three proteins similar to Porphyromonas gingivalis HmuY: Hemophore-like proteins differentially evolved to participate in heme acquisition in oral and gut microbiomes.
Faseb J., 37, 2023
|
|
8B6A
 
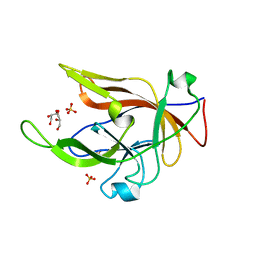 | |
8CCX
 
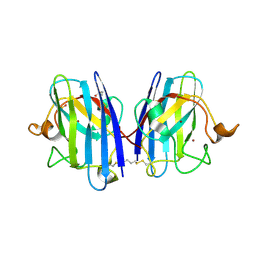 | | Human SOD1 in complex with S-XL6 cross-linker | | Descriptor: | COPPER (II) ION, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Hossain, A, Agar, J.N, Hasnain, S.S. | | Deposit date: | 2023-01-27 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Evaluating protein cross-linking as a therapeutic strategy to stabilize SOD1 variants in a mouse model of familial ALS.
Plos Biol., 22, 2024
|
|
2JLP
 
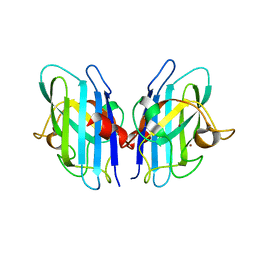 | | Crystal structure of human extracellular copper-zinc superoxide dismutase. | | Descriptor: | COPPER (II) ION, EXTRACELLULAR SUPEROXIDE DISMUTASE (CU-ZN), THIOCYANATE ION, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Marklund, S.L, Hasnain, S.S. | | Deposit date: | 2008-09-14 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Human Extracellular Copper-Zinc Superoxide Dismutase at 1.7 A Resolution: Insights Into Heparin and Collagen Binding.
J.Mol.Biol., 388, 2009
|
|
5G4P
 
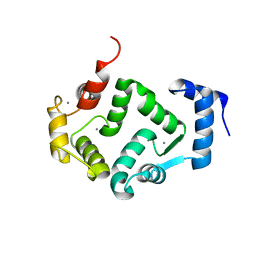 | | Crystal structure of human hippocalcin at 2.4 A resolution | | Descriptor: | CALCIUM ION, Neuron-specific calcium-binding protein hippocalcin | | Authors: | Antonyuk, S.V, Helassa, N, Lian, L.Y, Haynes, L.P, Burgoyne, R.D. | | Deposit date: | 2016-05-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Biophysical and functional characterization of hippocalcin mutants responsible for human dystonia.
Hum. Mol. Genet., 26, 2017
|
|
5G58
 
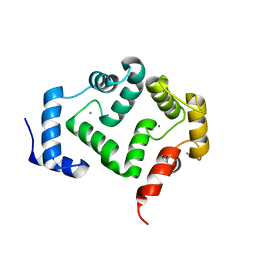 | | Crystal structure of A190T mutant of human hippocalcin AT 2.5 A resolution | | Descriptor: | CALCIUM ION, Neuron-specific calcium-binding protein hippocalcin | | Authors: | Antonyuk, S.V, Helassa, N, Lian, L.Y, Haynes, L.P, Burgoyne, R.D. | | Deposit date: | 2016-05-22 | | Release date: | 2017-05-03 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Biophysical and functional characterization of hippocalcin mutants responsible for human dystonia.
Hum. Mol. Genet., 26, 2017
|
|
3ZTZ
 
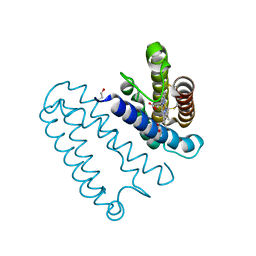 | | Cytochrome c prime from alcaligenes xylosoxidans: carbon monooxide bound L16G variant at 1.05 A resolution: unrestraint refinement | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-07-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZQV
 
 | |
3IXQ
 
 | | Structure of ribose 5-phosphate isomerase a from methanocaldococcus jannaschii | | Descriptor: | ACETATE ION, CHLORIDE ION, Ribose-5-phosphate isomerase A, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-04 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of an archaeal ribose-5-phosphate isomerase from Methanocaldococcus jannaschii (MJ1603).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3IWT
 
 | | Structure of hypothetical molybdenum cofactor biosynthesis protein B from Sulfolobus tokodaii | | Descriptor: | 178aa long hypothetical molybdenum cofactor biosynthesis protein B, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of hypothetical Mo-cofactor biosynthesis protein B (ST2315) from Sulfolobus tokodaii
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3KB6
 
 | | Crystal structure of D-Lactate dehydrogenase from aquifex aeolicus complexed with NAD and Lactic acid | | Descriptor: | D-lactate dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of D-lactate dehydrogenase from Aquifex aeolicus complexed with NAD(+) and lactic acid (or pyruvate).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1OBF
 
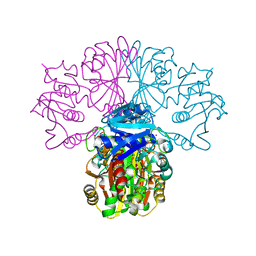 | | The crystal structure of Glyceraldehyde 3-phosphate Dehydrogenase from Alcaligenes xylosoxidans at 1.7A resolution. | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, POTASSIUM ION, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Eady, R.R, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Glyceraldehyde 3-Phosphate Dehydrogenase from Alcaligenes Xylosoxidans at 1.7 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OBG
 
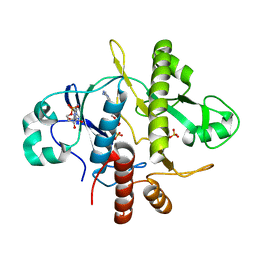 | | SAICAR-synthase complexed with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, PHOSPHORIBOSYLAMIDOIMIDAZOLE- SUCCINOCARBOXAMIDE SYNTHASE, ... | | Authors: | Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Urusova, D.V, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-Ray Structure of Saicar-Synthase Complexed with ATP
Kristallografiya, 46, 2001
|
|
1OBD
 
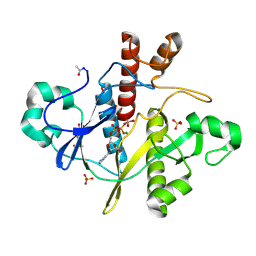 | | SAICAR-synthase complexed with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Urusova, D.V, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-Ray Structure of Saicar-Synthase Complexed with ATP
Kristallografiya, 46, 2001
|
|
2BWD
 
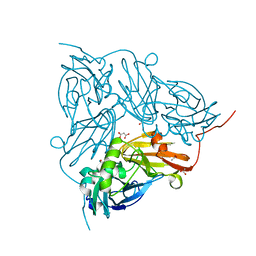 | | Atomic Resolution Structure of Achromobacter cycloclastes Cu Nitrite Reductase with Endogenously bound Nitrite and NO | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-13 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BW4
 
 | | Atomic Resolution Structure of Resting State of the Achromobacter cycloclastes Cu Nitrite Reductase | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-12 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BW5
 
 | | Atomic Resolution Structure of NO-bound Achromobacter cycloclastes Cu Nitrite Reductase | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-12 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BWI
 
 | | Atomic Resolution Structure of Nitrite -soaked Achromobacter cycloclastes Cu Nitrite Reductase | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-14 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3ZK4
 
 | |
3ZQY
 
 | | CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: CARBON MONOOXIDE BOUND L16A VARIANT AT 1.03 A RESOLUTION- NON-RESTRAINT REFINEMENT | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-06-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZTM
 
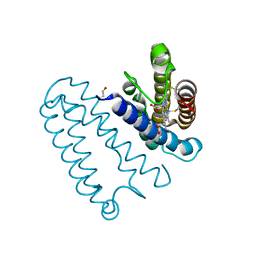 | | Cytochrome c prime from alcaligenes xylosoxidans: as isolated L16G variant at 0.9 A resolution: unrestraint refinement | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-07-11 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZBM
 
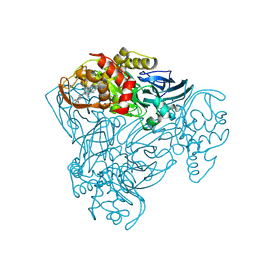 | | Structure of M92A variant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, HEME C | | Authors: | Antonyuk, S.V, Han, C, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2012-11-10 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structures of protein-protein complexes involved in electron transfer.
Nature, 496, 2013
|
|
3ZIY
 
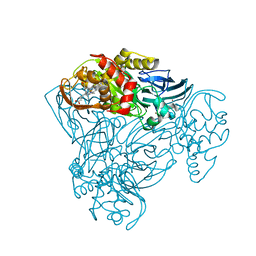 | | Structure of three-domain heme-Cu nitrite reductase from Ralstonia pickettii at 1.01 A resolution | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, HEME C | | Authors: | Antonyuk, S.V, Han, C, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2013-01-14 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structures of protein-protein complexes involved in electron transfer.
Nature, 496, 2013
|
|
4AX3
 
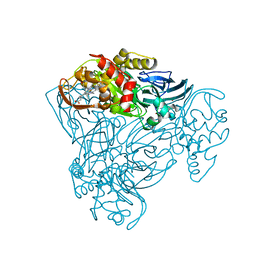 | | Structure of three-domain heme-Cu nitrite reductase from Ralstonia pickettii at 1.6 A resolution | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, HEME C | | Authors: | Antonyuk, S.V, Cong, H, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2012-06-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of protein-protein complexes involved in electron transfer.
Nature, 496, 2013
|
|
