1B94
 
 | | RESTRICTION ENDONUCLEASE ECORV WITH CALCIUM | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*TP*CP*TP*T)-3'), RESTRICTION ENDONUCLEASE ECORV | | Authors: | Thomas, M.P, Halford, S.E, Brady, R.L. | | Deposit date: | 1999-02-19 | | Release date: | 1999-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a mutational hot-spot in the EcoRV restriction endonuclease: a catalytic role for a main chain carbonyl group.
Nucleic Acids Res., 27, 1999
|
|
1B95
 
 | |
1B97
 
 | |
1B96
 
 | |
6BG0
 
 | | Caspase-3 Mutant - D9A,D28A,S150D | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, AZIDE ION, Caspase-3, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-27 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.125 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BFO
 
 | | Caspase-3 Mutant- T245D | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, AZIDE ION, Caspase-3, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BG4
 
 | | Caspase-3 Mutant- T152D | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, AZIDE ION, CHLORIDE ION, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-27 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BFK
 
 | | Caspase-3 Mutant- T245A | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, Caspase-3, SODIUM ION | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BGQ
 
 | | Caspase-3 Mutant - S150D | | Descriptor: | AZIDE ION, Ac-Asp-Glu-Val-Asp-CMK, CHLORIDE ION, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-29 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BGK
 
 | | Caspase-3 Mutant- D9A,D28A,T152D | | Descriptor: | ACE-ASP-GLU-VAL-ASP-0QE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-28 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BH9
 
 | | Caspase-3 Mutant - T152A | | Descriptor: | Ac-Asp-Glu-Val-Asp-CMK, CHLORIDE ION, Caspase-3, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-30 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
7ARE
 
 | | DNA origami pointer object v2 | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Thomas, M, Feigl, E, Kohler, F, Kube, M, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
6CWS
 
 | |
6BFJ
 
 | | Caspase-3 Mutant - T245D,S249D | | Descriptor: | AZIDE ION, Ac-Asp-Glu-Val-Asp-CMK, Caspase-3 | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BFL
 
 | | Caspase-3 Mutant- D9A,D28A,T245D | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, AZIDE ION, Caspase-3 | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BDV
 
 | | Crystal structure of Caspase 3 S150A | | Descriptor: | AZIDE ION, Acetyl-Asp-Glu-Val-Asp-CMK, CHLORIDE ION, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-24 | | Release date: | 2018-02-14 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BG1
 
 | | Caspase-3 Mutant - D9A,D28A,S150E | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, Caspase-3, SODIUM ION | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-27 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BGR
 
 | | Caspase-3 Mutant - S150E | | Descriptor: | AZIDE ION, Ac-Asp-Glu-Val-Asp-CMK, Caspase-3 | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-29 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BHA
 
 | | Caspase-3 Mutant - T152V | | Descriptor: | Ac-Asp-Glu-Val-Asp-CMK, Caspase-3 | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-30 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BGS
 
 | | Caspase-3 Mutant - S150Y | | Descriptor: | Ac-Asp-Glu-Val-Asp-CMK, Caspase-3 | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-29 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
1C9K
 
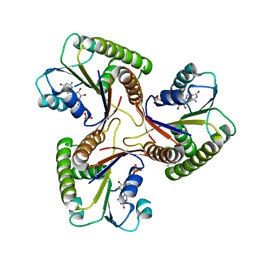 | | THE THREE DIMENSIONAL STRUCTURE OF ADENOSYLCOBINAMIDE KINASE/ ADENOSYLCOBINAMIDE PHOSPHATE GUALYLYLTRANSFERASE (COBU) COMPLEXED WITH GMP: EVIDENCE FOR A SUBSTRATE INDUCED TRANSFERASE ACTIVE SITE | | Descriptor: | ADENOSYLCOBINAMIDE KINASE, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thompson, T.B, Thomas, M.G, Esclante-Semerena, J.C, Rayment, I. | | Deposit date: | 1999-08-02 | | Release date: | 1999-08-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase (CobU) complexed with GMP: evidence for a substrate-induced transferase active site.
Biochemistry, 38, 1999
|
|
1CBU
 
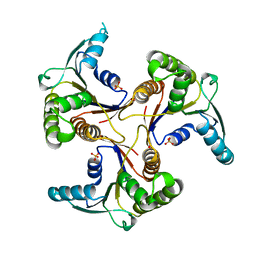 | | ADENOSYLCOBINAMIDE KINASE/ADENOSYLCOBINAMIDE PHOSPHATE GUANYLYLTRANSFERASE (COBU) FROM SALMONELLA TYPHIMURIUM | | Descriptor: | ADENOSYLCOBINAMIDE KINASE/ADENOSYLCOBINAMIDE PHOSPHATE GUANYLYLTRANSFERASE, SULFATE ION | | Authors: | Thompson, T.B, Thomas, M.G, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 1998-03-12 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase from Salmonella typhimurium determined to 2.3 A resolution,.
Biochemistry, 37, 1998
|
|
6XVA
 
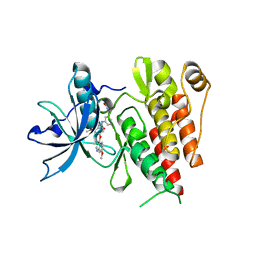 | | Crystal structure of the kinase domain of human c-KIT in complex with a type-II inhibitor bearing an acrylamide | | Descriptor: | Mast/stem cell growth factor receptor Kit,Mast/stem cell growth factor receptor Kit, ~{N}-[[3-[2-[3-methoxy-5-(7-methoxyquinolin-4-yl)oxy-pyridin-2-yl]ethanoylamino]-5-methyl-phenyl]methyl]propanamide | | Authors: | Schimpl, M, McAuley, K, Hoyt, E.A, Thomas, M, Bodnarchuk, M.S, Lewis, H.J, Barratt, D, Deery, M.J, Ogg, D.J, Bernardes, G.J.L, Ward, R.A, Kettle, J.G, Waring, M.J. | | Deposit date: | 2020-01-21 | | Release date: | 2020-05-27 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alkynyl Benzoxazines and Dihydroquinazolines as Cysteine Targeting Covalent Warheads and Their Application in Identification of Selective Irreversible Kinase Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6XVB
 
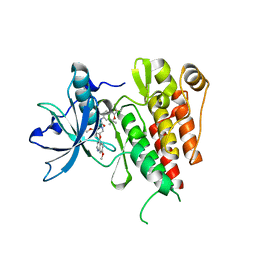 | | Crystal structure of the kinase domain of human c-KIT with a cyclic imidate inhibitor covalently bound to Cys788 | | Descriptor: | Mast/stem cell growth factor receptor Kit,Mast/stem cell growth factor receptor Kit, ~{N}-(4,4-dimethyl-2-propyl-3,1-benzoxazin-6-yl)-2-[3-methoxy-5-(7-methoxyquinolin-4-yl)oxy-pyridin-2-yl]ethanamide | | Authors: | Schimpl, M, McAuley, K, Hoyt, E.A, Thomas, M, Bodnarchuk, M.S, Lewis, H.J, Barratt, D, Deery, M.J, Ogg, D.J, Bernardes, G.J.L, Ward, R.A, Kettle, J.G, Waring, M.J. | | Deposit date: | 2020-01-21 | | Release date: | 2020-05-27 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Alkynyl Benzoxazines and Dihydroquinazolines as Cysteine Targeting Covalent Warheads and Their Application in Identification of Selective Irreversible Kinase Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | Authors: | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-07 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
