3U3G
 
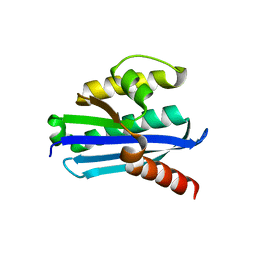 | | Structure of LC11-RNase H1 Isolated from Compost by Metagenomic Approach: Insight into the Structural Bases for Unusual Enzymatic Properties of Sto-RNase H1 | | Descriptor: | CHLORIDE ION, Ribonuclease H, UNKNOWN LIGAND | | Authors: | Nguyen, T.N, Angkawidjaja, C, Kanaya, E, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-10-05 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Activity, stability, and structure of metagenome-derived LC11-RNase H1, a homolog of Sulfolobus tokodaii RNase H1
Protein Sci., 21, 2012
|
|
4Q0B
 
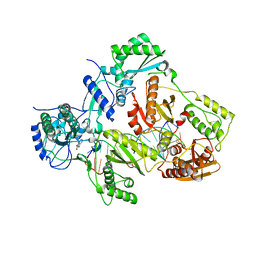 | | Crystal structure of HIV-1 reverse transcriptase in complex with gap-RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*G)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-04-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
4PWD
 
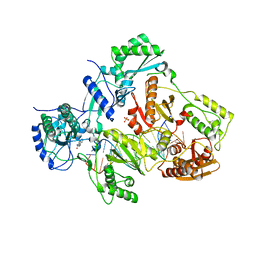 | | Crystal structure of HIV-1 reverse transcriptase in complex with bulge-RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*AP*CP*AP*GP*GP*GP*AP*CP*UP*GP*U)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-03-19 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
7WL0
 
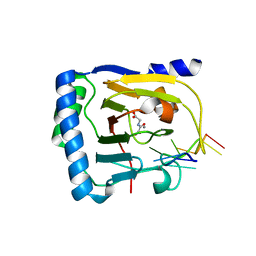 | | Crystal structure of human ALKBH5 in complex with N-oxalylglycine (NOG) and m6A-containing ssRNA | | Descriptor: | FORMIC ACID, MANGANESE (II) ION, N-OXALYLGLYCINE, ... | | Authors: | Kaur, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms of substrate recognition and N6-methyladenosine demethylation revealed by crystal structures of ALKBH5-RNA complexes.
Nucleic Acids Res., 50, 2022
|
|
1FIR
 
 | |
7V4G
 
 | | Crystal structure of human ALKBH5 in complex with m6A-containing ssRNA | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RNA (5'-R(P*GP*GP*(6MZ)P*C)-3'), ... | | Authors: | Kaur, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | Deposit date: | 2021-08-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanisms of substrate recognition and N6-methyladenosine demethylation revealed by crystal structures of ALKBH5-RNA complexes.
Nucleic Acids Res., 50, 2022
|
|
8H53
 
 | | Human asparaginyl-tRNA synthetase in complex with asparagine-AMP | | Descriptor: | 4-AMINO-1,4-DIOXOBUTAN-2-AMINIUM ADENOSINE-5'-MONOPHOSPHATE, Asparagine--tRNA ligase, cytoplasmic, ... | | Authors: | Park, J.S, Han, B.W. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
7WKV
 
 | | Crystal structure of human ALKBH5 in complex with 2-oxoglutarate (2OG) and m6A-containing ssRNA | | Descriptor: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, RNA (5'-R(P*GP*GP*(6MZ)P*C)-3'), ... | | Authors: | Kaur, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | Deposit date: | 2022-01-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanisms of substrate recognition and N6-methyladenosine demethylation revealed by crystal structures of ALKBH5-RNA complexes.
Nucleic Acids Res., 50, 2022
|
|
3J5S
 
 | |
1MMS
 
 | | Crystal structure of the ribosomal PROTEIN L11-RNA complex | | Descriptor: | 23S RIBOSOMAL RNA, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Wimberly, B.T, Guymon, R, Mccutcheon, J.P, White, S.W, Ramakrishnan, V. | | Deposit date: | 1999-04-14 | | Release date: | 2000-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A detailed view of a ribosomal active site: the structure of the L11-RNA complex.
Cell(Cambridge,Mass.), 97, 1999
|
|
4WLS
 
 | | Crystal structure of the metal-free (repressor) form of E. Coli CUER, a copper efflux regulator, bound to COPA promoter DNA | | Descriptor: | COPA PROMOTER DNA NON-TEMPLATE STRAND, COPA PROMOTER DNA NON-TEMPLATE STRAND (ALTERNATE CONFORMATION), COPA PROMOTER DNA TEMPLATE STRAND, ... | | Authors: | Philips, S.J, Canalizo-Hernandez, M, Mondragon, A, O'Halloran, T.V. | | Deposit date: | 2014-10-08 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Allosteric transcriptional regulation via changes in the overall topology of the core promoter.
Science, 349, 2015
|
|
3WQY
 
 | | Crystal structure of Archaeoglobus fulgidus alanyl-tRNA synthetase in complex with wild-type tRNA(Ala) having G3.U70 | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, Alanine--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Naganuma, M, Sekine, S, Yokoyama, S. | | Deposit date: | 2014-02-05 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The selective tRNA aminoacylation mechanism based on a single G.U pair
Nature, 510, 2014
|
|
3WQZ
 
 | | Crystal structure of Archaeoglobus fulgidus alanyl-tRNA synthetase in complex with a tRNA(Ala) variant having A3.U70 | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, Alanine--tRNA ligase, RNA (75-MER), ... | | Authors: | Naganuma, M, Sekine, S, Yokoyama, S. | | Deposit date: | 2014-02-05 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.489 Å) | | Cite: | The selective tRNA aminoacylation mechanism based on a single G.U pair
Nature, 510, 2014
|
|
4LWQ
 
 | | Crystal structure of native peptidyl t-RNA hydrolase from Acinetobacter baumannii at 1.38A resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Kaushik, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-28 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of native peptidyl t-RNA hydrolase from Acinetobacter baumannii at 1.38A resolution
To be Published
|
|
7Q4L
 
 | |
8R8R
 
 | | Cryo-EM structure of the human mPSF with PAPOA C-terminus peptide (PAPOAc) | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*AP*AP*UP*AP*AP*A)-3'), ... | | Authors: | Todesca, S, Sandmeir, F, Keidel, A, Conti, E. | | Deposit date: | 2023-11-29 | | Release date: | 2024-04-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis of human poly(A) polymerase recruitment by mPSF.
Rna, 30, 2024
|
|
7DPI
 
 | | Plasmodium falciparum cytoplasmic Phenylalanyl-tRNA synthetase in complex with BRD7929 | | Descriptor: | (8R,9S,10S)-10-[(dimethylamino)methyl]-N-(4-methoxyphenyl)-9-[4-(2-phenylethynyl)phenyl]-1,6-diazabicyclo[6.2.0]decane-6-carboxamide, MAGNESIUM ION, Phenylalanine--tRNA ligase, ... | | Authors: | Manmohan, S, Malhotra, N, Harlos, K, Manickam, Y, Sharma, A. | | Deposit date: | 2020-12-19 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.597 Å) | | Cite: | Inhibition of Plasmodium falciparum phenylalanine tRNA synthetase provides opportunity for antimalarial drug development.
Structure, 30, 2022
|
|
7TZB
 
 | | Crystal structure of the human mitochondrial seryl-tRNA synthetase (mt SerRS) bound with a seryl-adenylate analogue | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Serine--tRNA ligase, mitochondrial | | Authors: | Kuhle, B, Hirschi, M, Doerfel, L, Lander, G, Schimmel, P. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
7E44
 
 | | Crystal structure of NudC complexed with dpCoA | | Descriptor: | DEPHOSPHO COENZYME A, NADH pyrophosphatase, ZINC ION | | Authors: | Zhou, W, Guan, Z.Y, Yin, P, Zhang, D.L. | | Deposit date: | 2021-02-10 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into dpCoA-RNA decapping by NudC.
Rna Biol., 18, 2021
|
|
7ULZ
 
 | |
5AH5
 
 | | Crystal structure of the ternary complex of Agrobacterium radiobacter K84 agnB2 LeuRS-tRNA-LeuAMS | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, LEUCINE--TRNA LIGASE, MANGANESE (II) ION, ... | | Authors: | Palencia, A, Chopra, S, Virus, C, Schulwitz, S, Temple, B.R, Cusack, S, Reader, J.S. | | Deposit date: | 2015-02-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Antibiotic Self-Immunity tRNA Synthetase in Plant Tumour Biocontrol Agent.
Nat.Commun., 7, 2016
|
|
2BH2
 
 | | Crystal Structure of E. coli 5-methyluridine methyltransferase RumA in complex with ribosomal RNA substrate and S-adenosylhomocysteine. | | Descriptor: | 23S RIBOSOMAL RNA 1932-1968, 23S RRNA (URACIL-5-)-METHYLTRANSFERASE RUMA, IRON/SULFUR CLUSTER, ... | | Authors: | Lee, T.T, Agarwalla, S, Stroud, R.M. | | Deposit date: | 2005-01-06 | | Release date: | 2005-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Unique RNA Fold in the Ruma-RNA-Cofactor Ternary Complex Contributes to Substrate Selectivity and Enzymatic Function
Cell(Cambridge,Mass.), 120, 2005
|
|
8Y04
 
 | | Crystal structure of LbCas12a in complex with crRNA and 6nt target DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*TP*TP*TP*AP*TP*T)-3'), DNA (5'-D(P*GP*AP*TP*GP*CP*GP*TP*AP*AP*AP*GP*GP*AP*CP*G)-3'), LbCas12a, ... | | Authors: | Lin, X, Chen, J, Liu, L. | | Deposit date: | 2024-01-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | DNA target binding-induced pre-crRNA processing in type II and V CRISPR-Cas systems.
Nucleic Acids Res., 53, 2025
|
|
8Y0A
 
 | | Crystal structure of LbCas12a in complex with crRNA and 18nt target DNA | | Descriptor: | DNA (27-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*TP*TP*AP*TP*T)-3'), LbCas12a, ... | | Authors: | Lin, X, Chen, J, Liu, L. | | Deposit date: | 2024-01-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | DNA target binding-induced pre-crRNA processing in type II and V CRISPR-Cas systems.
Nucleic Acids Res., 53, 2025
|
|
8Y06
 
 | | Crystal structure of LbCas12a in complex with crRNA and 12nt target DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*TP*TP*TP*AP*TP*T)-3'), DNA (5'-D(P*TP*TP*AP*CP*TP*GP*GP*AP*TP*GP*CP*GP*TP*AP*AP*AP*GP*GP*AP*CP*G)-3'), LbCas12a, ... | | Authors: | Lin, X, Chen, J, Liu, L. | | Deposit date: | 2024-01-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.99 Å) | | Cite: | DNA target binding-induced pre-crRNA processing in type II and V CRISPR-Cas systems.
Nucleic Acids Res., 53, 2025
|
|
