5JGI
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with M45 compound | | Descriptor: | N-ALPHA-L-ACETYL-ARGININE, Neuropilin-1 | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
5JGQ
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-7 ligand. | | Descriptor: | DIMETHYL SULFOXIDE, Neuropilin-1, N~2~-(benzenecarbonyl)-L-arginine | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
6OY1
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-26 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-ethylbutyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXO
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-91 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[4-(hydroxymethyl)phenyl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXP
 
 | | HIV-1 Protease NL4-3 WT in Complex with UMass3 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)propyl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXS
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR-76 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[({4-[(1R)-1-hydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXX
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-18 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
3UX9
 
 | | Structural insights into a human anti-IFN antibody exerting therapeutic potential for systemic lupus erythematosus | | Descriptor: | Interferon alpha-1/13, ScFv antibody | | Authors: | Ouyang, S, Zhao, L.X, Liang, W, Shaw, N, Liu, Z.-J, Liang, M.-F. | | Deposit date: | 2011-12-04 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into a human anti-IFN antibody exerting therapeutic potential for systemic lupus erythematosus
J.Mol.Med., 2012
|
|
6OXQ
 
 | | HIV-1 Protease NL4-3 WT in Complex with UMass8 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-1-benzyl-3-[(2-ethylbutyl){[4-(hydroxymethyl)phenyl]sulfonyl}amino]-2-hydroxypropyl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXV
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR-85 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[(2-ethylbutyl)({4-[(1S)-1-hydroxyethyl]phenyl}sulfonyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXY
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-19 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
5IJR
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-1 ligand. | | Descriptor: | DIMETHYL SULFOXIDE, L-HOMOARGININE, Neuropilin-1 | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-03-02 | | Release date: | 2017-03-29 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
5KIT
 
 | |
6OXT
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR-84 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{({4-[(1S)-1-hydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OY0
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-21 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
5J1X
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-5 ligand. | | Descriptor: | DIMETHYL SULFOXIDE, Neuropilin-1, N~2~-(tert-butoxycarbonyl)-L-arginine | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-03-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
5JHK
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-6 ligand. | | Descriptor: | N-(benzenecarbonyl)glycyl-L-arginine, Neuropilin-1 | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-04-21 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
4AJC
 
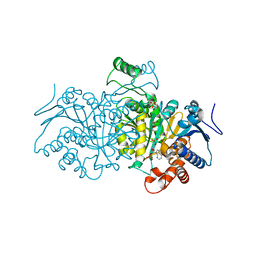 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with alpha-ketoglutarate, calcium(II) and adenine nucleotide phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, ADENOSINE-2'-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJB
 
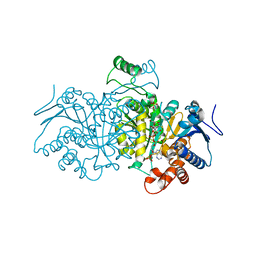 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with Isocitrate, magnesium(II) and thioNADP | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJS
 
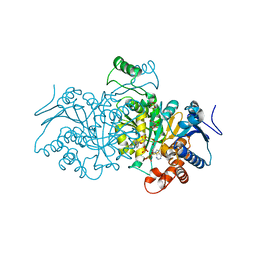 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with isocitrate, magnesium(II), Adenosine 2',5'-biphosphate and ribosylnicotinamide-5'-phosphate | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ISOCITRATE DEHYDROGENASE [NADP], ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJR
 
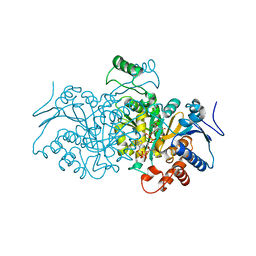 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with alpha-ketoglutarate, magnesium(II) and NADPH - The product complex | | Descriptor: | 2-OXOGLUTARIC ACID, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ISOCITRATE DEHYDROGENASE [NADP], ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.687 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJ3
 
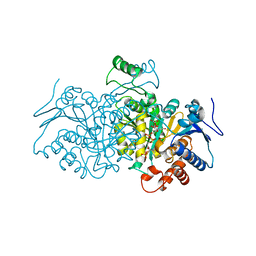 | | 3D structure of E. coli Isocitrate Dehydrogenase in complex with Isocitrate, calcium(II) and NADP - The pseudo-Michaelis complex | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, NADP ISOCITRATE DEHYDROGENASE, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJA
 
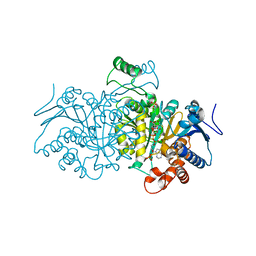 | | 3D structure of E. coli Isocitrate Dehydrogenase in complex with Isocitrate, calcium(II) and thioNADP | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, CALCIUM ION, ISOCITRIC ACID, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
7VSG
 
 | |
7VSH
 
 | |
