4LH5
 
 | | Dual inhibition of HIV-1 replication by Integrase-LEDGF allosteric inhibitors is predominant at post-integration stage during virus production rather than at integration | | Descriptor: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, Integrase, MAGNESIUM ION | | Authors: | Ruff, M, Levy, N, Eiler, S. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage.
Retrovirology, 10, 2013
|
|
1JJ9
 
 | | Crystal Structure of MMP8-Barbiturate Complex Reveals Mechanism for Collagen Substrate Recognition | | Descriptor: | 2-HYDROXY-5-[4-(2-HYDROXY-ETHYL)-PIPERIDIN-1-YL]-5-PHENYL-1H-PYRIMIDINE-4,6-DIONE, CALCIUM ION, Matrix Metalloproteinase 8, ... | | Authors: | Brandstetter, H, Grams, F, Glitz, D, Lang, A, Huber, R, Bode, W, Krell, H.-W, Engh, R.A. | | Deposit date: | 2001-07-04 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 1.8-A crystal structure of a matrix metalloproteinase 8-barbiturate inhibitor complex reveals a previously unobserved mechanism for collagenase substrate recognition.
J.Biol.Chem., 276, 2001
|
|
7ZM8
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
4LM3
 
 | | Crystal structure of PDE10A2 with fragment ZT464 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2,3-dihydro-1,4-benzodioxin-6-yl)ethanone, NICKEL (II) ION, ... | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4MW0
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea (Chem 1392) | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
1T3G
 
 | |
4C2J
 
 | | Crystal structure of human mitochondrial 3-ketoacyl-CoA thiolase in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, 3-KETOACYL-COA THIOLASE, MITOCHONDRIAL, ... | | Authors: | Kiema, T.-R, Harijan, R.K, Wierenga, R.K. | | Deposit date: | 2013-08-19 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Mitochondrial 3-Ketoacyl-Coa Thiolase (T1): Insight Into the Reaction Mechanism of its Thiolase and Thioesterase Activities
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C6H
 
 | | Haloalkane dehalogenase with 1-hexanol | | Descriptor: | CHLORIDE ION, HALOALKANE DEHALOGENASE, HEXAN-1-OL, ... | | Authors: | Novak, H.R, Sayer, C, Isupov, M, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae.
FEBS Lett., 588, 2014
|
|
1JP5
 
 | | Crystal structure of the single-chain Fv fragment 1696 in complex with the epitope peptide corresponding to N-terminus of HIV-1 protease | | Descriptor: | epitope peptide corresponding to N-terminus of HIV-1 protease, single-chain Fv fragment 1696 | | Authors: | Rezacova, P, Lescar, J, Brynda, J, Fabry, M, Horejsi, M, Sedlacek, J, Bentley, G.A. | | Deposit date: | 2001-08-01 | | Release date: | 2001-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of HIV-1 and HIV-2 protease inhibition by a monoclonal antibody.
Structure, 9, 2001
|
|
3VYF
 
 | | Human renin in complex with inhibitor 9 | | Descriptor: | (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]-N-(2,6-dimethylheptan-4-yl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2012-09-24 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and discovery of new (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]piperidine-3-carboxamides as potent renin inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6N9D
 
 | | Complex of tissue inhibitor of metalloproteinases-1 (TIMP-1) mutant (L34G/L133P/L151C/G154A) with matrix metalloproteinase-3 catalytic domain (MMP-3cd) | | Descriptor: | CALCIUM ION, Metalloproteinase inhibitor 1, Stromelysin-1, ... | | Authors: | Raeeszadeh-Sarmazdeh, M, Radisky, E.S, Sankaran, B. | | Deposit date: | 2018-12-03 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Directed evolution of the metalloproteinase inhibitor TIMP-1 reveals that its N- and C-terminal domains cooperate in matrix metalloproteinase recognition.
J.Biol.Chem., 294, 2019
|
|
1B8N
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-1-(S)-(9-DEAZAGUANIN-9-YL)-D-RIBITOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fedorov, A.A, Kicska, G.A, Fedorov, E.V, Strokopytov, B.V, Tyler, P.C, Furneaux, R.H, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic dissection of the hydrogen bond network for transition-state analogue binding to purine nucleoside phosphorylase
Biochemistry, 41, 2002
|
|
3BL4
 
 | |
4MUL
 
 | |
1BC8
 
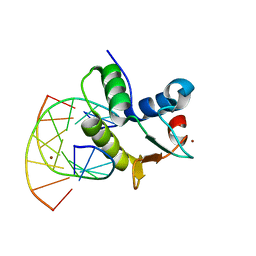 | | STRUCTURES OF SAP-1 BOUND TO DNA SEQUENCES FROM THE E74 AND C-FOS PROMOTERS PROVIDE INSIGHTS INTO HOW ETS PROTEINS DISCRIMINATE BETWEEN RELATED DNA TARGETS | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*TP*CP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*GP*GP*AP*AP*GP*T)-3'), PROTEIN (SAP-1 ETS DOMAIN), ... | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 1998-05-05 | | Release date: | 1998-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins.
Mol.Cell, 2, 1998
|
|
1B8O
 
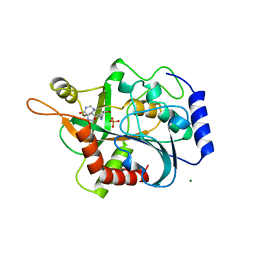 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fedorov, A.A, Kicska, G.A, Fedorov, E.V, Shi, W, Tyler, P.C, Furneaux, R.H, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis.
Biochemistry, 40, 2001
|
|
3CIV
 
 | |
4C4J
 
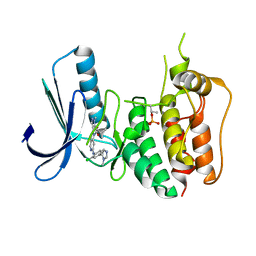 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE TTK, tert-butyl 6-{[2-chloro-4-(1-methyl-1H-imidazol-5-yl)phenyl]amino}-2-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridine-1-carboxylate | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
3BUH
 
 | | BACE-1 complexed with compound 4 | | Descriptor: | 4-(2-aminoethyl)-2-cyclohexylphenol, beta-secretase 1 | | Authors: | Kuglstatter, A, Hennig, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tyramine fragment binding to BACE-1
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1FN6
 
 | | CRYSTAL STRUCTURE OF PORCINE BETA TRYPSIN WITH 0.1% POLYDOCANOL | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, METHANOL, ... | | Authors: | Deepthi, S, Johnson, A, Pattabhi, V. | | Deposit date: | 2000-08-21 | | Release date: | 2000-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of porcine beta-trypsin-detergent complexes: the stabilization of proteins through hydrophilic binding of polydocanol.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3BWW
 
 | |
4BP0
 
 | | Crystal structure of the closed form of Pseudomonas aeruginosa SPM-1 | | Descriptor: | CHLORIDE ION, GLYCEROL, METALLO-B-LACTAMASE, ... | | Authors: | McDonough, M.A, Brem, J, Schofield, C.J. | | Deposit date: | 2013-05-22 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Studying the active-site loop movement of the Sao Paolo metallo-beta-lactamase-1
Chem Sci, 6, 2015
|
|
3CMH
 
 | | SYNTHETIC LINEAR TRUNCATED ENDOTHELIN-1 AGONIST | | Descriptor: | PROTEIN (ENDOTHELIN-1) | | Authors: | Hewage, C.M, Jiang, L, Parkinson, J.A, Ramage, R, Sadler, I.H. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel ETB receptor selective agonist ET1-21 [Cys(Acm)1,15, Aib3,11, Leu7] by nuclear magnetic resonance spectroscopy and molecular modelling.
J.Pept.Res., 53, 1999
|
|
5UB5
 
 | | human POGLUT1 in complex with human Notch1 EGF12 S458T mutant and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurogenic locus notch homolog protein 1, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-12-20 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
4HA5
 
 | | Structure of BACE Bound to (S)-3-(5-(2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl)thiophen-3-yl)benzonitrile | | Descriptor: | 3-{5-[(2E,4S)-2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
