8T1O
 
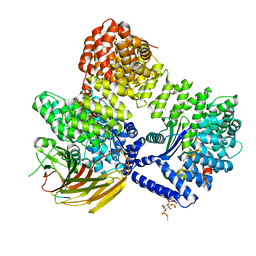 | | AP2 bound to MSP2N2 nanodisc with Tgn38 cargo peptide; composite map | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Cannon, K.S, Reta, S. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lipid nanodiscs as a template for high-resolution cryo-EM structures of peripheral membrane proteins.
J.Struct.Biol., 215, 2023
|
|
7RWC
 
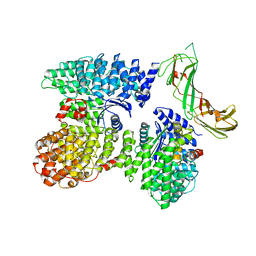 | | AP2 bound to the APA domain of SGIP and heparin; partial signal subtraction and symmetry expansion | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Hollopeter, G, Partlow, E.A. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RWA
 
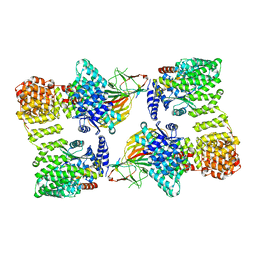 | | AP2 bound to heparin and Tgn38 tyrosine cargo peptide | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Hollopeter, G, Partlow, E.A. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RWB
 
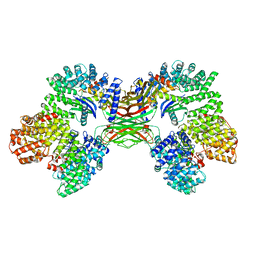 | | AP2 bound to the APA domain of SGIP in the presence of heparin | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Hollopeter, G, Partlow, E.A. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RW9
 
 | | AP2 bound to heparin in the bowl conformation | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Hollopeter, G, Partlow, E.A. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RW8
 
 | | AP2 bound to heparin in the closed conformation | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Hollopeter, G, Partlow, E.A. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1JHS
 
 | | Protein Mog1 E65A mutant | | Descriptor: | MOG1 PROTEIN | | Authors: | Baker, R.P, Harreman, M.T, Ecclestone, J.F, Corbett, A.H, Stewart, M. | | Deposit date: | 2001-06-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction between Ran and Mog1 is required for efficient nuclear protein import
J.Biol.Chem., 276, 2001
|
|
5BUZ
 
 | |
5BV0
 
 | |
5BV1
 
 | |
4KMO
 
 | | Crystal Structure of the Vps33-Vps16 HOPS subcomplex from Chaetomium thermophilum | | Descriptor: | Putative vacuolar protein sorting-associated protein, SULFATE ION, Small conjugating protein ligase-like protein | | Authors: | Baker, R.W, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2013-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Sec1/Munc18 (SM) Protein Vps33, Alone and Bound to the Homotypic Fusion and Vacuolar Protein Sorting (HOPS) Subunit Vps16*
Plos One, 8, 2013
|
|
4JC8
 
 | |
1A0P
 
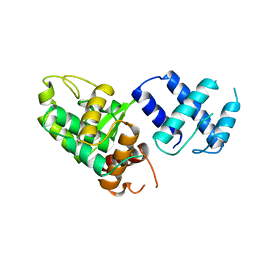 | | SITE-SPECIFIC RECOMBINASE, XERD | | Descriptor: | SITE-SPECIFIC RECOMBINASE XERD | | Authors: | Subramanya, H.S, Arciszewska, L.K, Baker, R.A, Bird, L.E, Sherratt, D.J, Wigley, D.B. | | Deposit date: | 1997-12-05 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the site-specific recombinase, XerD.
EMBO J., 16, 1997
|
|
8G45
 
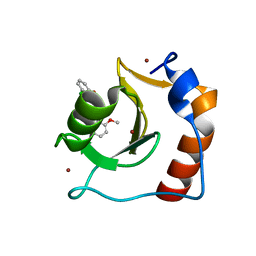 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with SGC-UBD253 chemical probe | | Descriptor: | 3-[8-chloro-3-(2-{[(2-methoxyphenyl)methyl]amino}-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl]propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G43
 
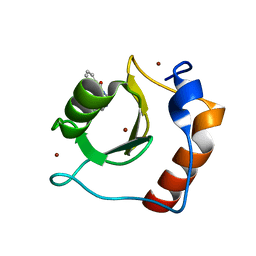 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(methylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(methylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G44
 
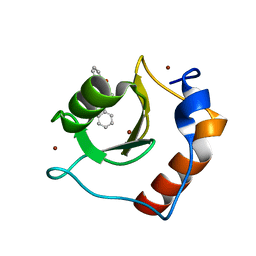 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(benzylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(benzylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
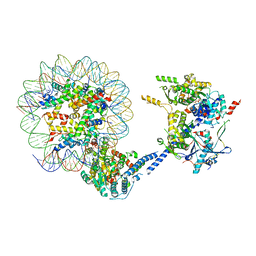
6VZ4
 
 | |
8CUK
 
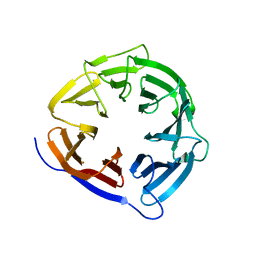 | |
6VZG
 
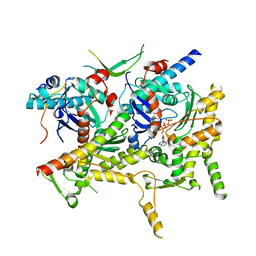 | | Cryo-EM structure of Sth1-Arp7-Arp9-Rtt102 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-like protein ARP9, Actin-related protein 7, ... | | Authors: | Leschziner, A.E, Baker, R.W. | | Deposit date: | 2020-02-28 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into assembly and function of the RSC chromatin remodeling complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6V9I
 
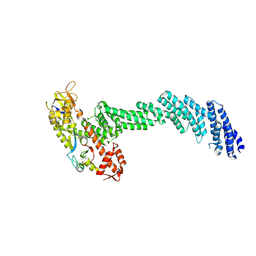 | | cryo-EM structure of Cullin5 bound to RING-box protein 2 (Cul5-Rbx2) | | Descriptor: | Immunoglobulin G-binding protein G,Cullin-5, RING-box protein 2, ZINC ION | | Authors: | Komives, E.A, Lumpkin, R.J, Baker, R.W, Leschziner, A.E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-04-29 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structure and dynamics of the ASB9 CUL-RING E3 Ligase.
Nat Commun, 11, 2020
|
|
6V9H
 
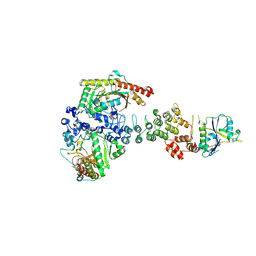 | | Ankyrin repeat and SOCS-box protein 9 (ASB9), ElonginB (ELOB), and ElonginC (ELOC) bound to its substrate Brain-type Creatine Kinase (CKB) | | Descriptor: | Ankyrin repeat and SOCS box protein 9, Creatine kinase B-type, Elongin-B, ... | | Authors: | Komives, E.A, Lumpkin, R.J, Baker, R.W, Leschziner, A.E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-04-29 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and dynamics of the ASB9 CUL-RING E3 Ligase.
Nat Commun, 11, 2020
|
|
1EQ6
 
 | |
6OXL
 
 | | CRYO-EM STRUCTURE OF PHOSPHORYLATED AP-2 (mu E302K) BOUND TO NECAP IN THE PRESENCE OF SS DNA | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Partlow, E.A, Baker, R.W, Beacham, G.M, Chappie, J, Leschziner, A.E, Hollopeter, G. | | Deposit date: | 2019-05-13 | | Release date: | 2019-09-11 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A structural mechanism for phosphorylation-dependent inactivation of the AP2 complex.
Elife, 8, 2019
|
|
6OWO
 
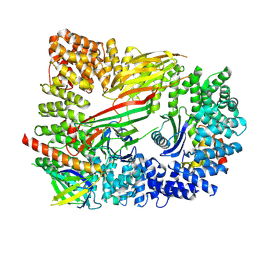 | | CRYO-EM STRUCTURE OF PHOSPHORYLATED AP-2 CORE BOUND TO NECAP | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Partlow, E.A, Baker, R.W, Beacham, G.M, Chappie, J.S, Leschziner, A.E, Hollopeter, G. | | Deposit date: | 2019-05-10 | | Release date: | 2019-09-11 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A structural mechanism for phosphorylation-dependent inactivation of the AP2 complex.
Elife, 8, 2019
|
|
4NJN
 
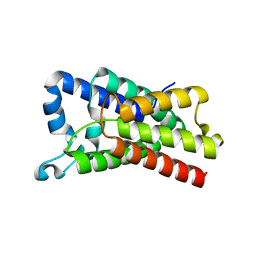 | | Crystal Structure of E.coli GlpG at pH 4.5 | | Descriptor: | Rhomboid protease GlpG | | Authors: | Dickey, S.W, Baker, R.P, Cho, S, Urban, S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Proteolysis inside the Membrane Is a Rate-Governed Reaction Not Driven by Substrate Affinity.
Cell(Cambridge,Mass.), 155, 2013
|
|
