9CF0
 
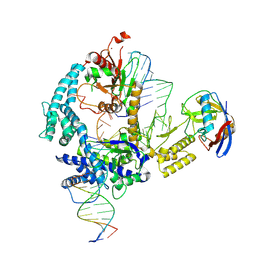 | | Parasitella parasitica Fanzor (PpFz) State 1 | | Descriptor: | DNA non-target strand, DNA target strand, Maltose/maltodextrin-binding periplasmic protein,Parasitella parasitica Fanzor 1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEX
 
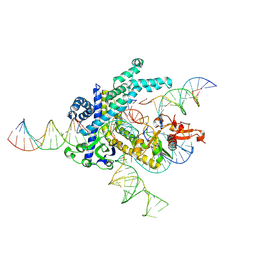 | | Spizellomyces punctatus Fanzor (SpuFz) State 4 | | Descriptor: | DNA (29-MER), DNA (5'-D(*(MG)*(MG)P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CET
 
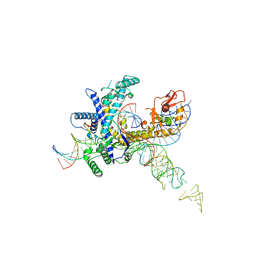 | | Guillardia theta Fanzor (GtFz) State 3 | | Descriptor: | DNA (28-MER), DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF1
 
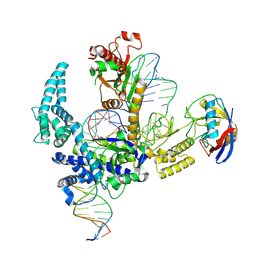 | | Parasitella parasitica Fanzor (PpFz) State 2 | | Descriptor: | DNA non-target strand, DNA target strand, Maltose/maltodextrin-binding periplasmic protein,Parasitella parasitica Fanzor 1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CES
 
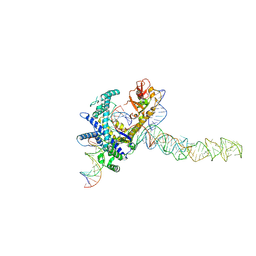 | | Guillardia theta Fanzor (GtFz) State 2 | | Descriptor: | DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*GP*GP*GP*GP*CP*CP*TP*TP*TP*AP*AP*G)-3'), Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9B1D
 
 | |
9B1E
 
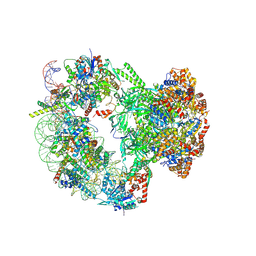 | |
7JTR
 
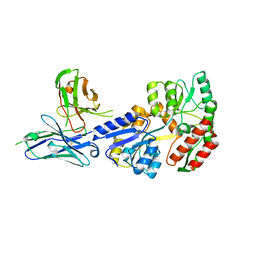 | | Complex of maltose-binding protein (MBP) with single-chain Fv (scFv) | | Descriptor: | CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Loll, P.J. | | Deposit date: | 2020-08-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A useful epitope tag derived from maltose binding protein.
Protein Sci., 30, 2021
|
|
7JHG
 
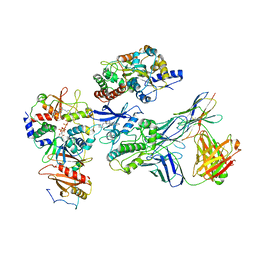 | | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compound C) and Fab-nanobody | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Murkherjee, S, Zhou, X.E, Xu, T.H, Xu, H.E, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7JIJ
 
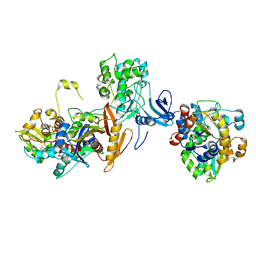 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Powell, K, Xu, T, Brunzelle, J.S, Xu, H.X, Melcher, K. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7JHH
 
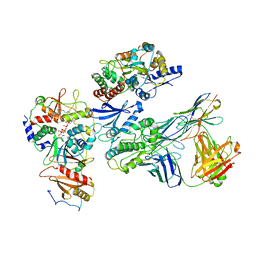 | | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Fab and nanobody | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Murkherjee, S, Zhou, X.E, Xu, T.H, Xu, H.E, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7K48
 
 | | Structure of NavAb/Nav1.7-VS2A chimera trapped in the resting state by tarantula toxin m3-Huwentoxin-IV | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Ion transport protein,Sodium channel protein type 9 subunit alpha chimera, Mu-theraphotoxin-Hs2a | | Authors: | Wisedchaisri, G, Tonggu, L, Gamal El-Din, T.M, McCord, E, Zheng, N, Catterall, W.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for High-Affinity Trapping of the Na V 1.7 Channel in Its Resting State by Tarantula Toxin.
Mol.Cell, 81, 2021
|
|
7KD4
 
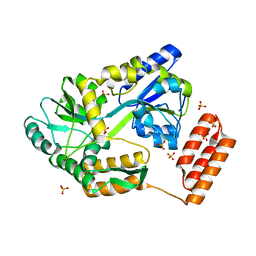 | |
7KD5
 
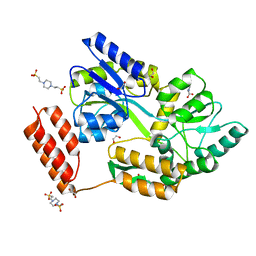 | | Structure of the C-terminal domain of the Menangle virus phosphoprotein (residues 329 -388), fused to MBP. Space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, Maltodextrin-binding protein and Phosphoprotein fusion protein, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Webby, M.N, Kingston, R.L. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural Analysis of the Menangle Virus P Protein Reveals a Soft Boundary between Ordered and Disordered Regions.
Viruses, 13, 2021
|
|
5TIB
 
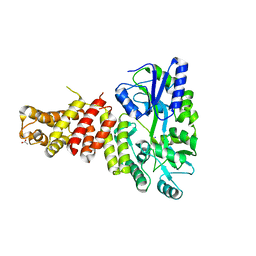 | | Gasdermin-B C-terminal domain containing the polymorphism residues Arg299:Ser306 fused to maltose binding protein | | Descriptor: | ACETATE ION, SODIUM ION, Sugar ABC transporter substrate-binding protein,Gasdermin-B, ... | | Authors: | Chao, K, Herzberg, O. | | Deposit date: | 2016-10-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human Gasdermin-B and disease: Sulfatide Binding, Caspase cleavage, and Structural impact of Asthma- and IBS-Associated Polymorphism
Proc.Natl.Acad.Sci.Usa, 2017
|
|
5UB5
 
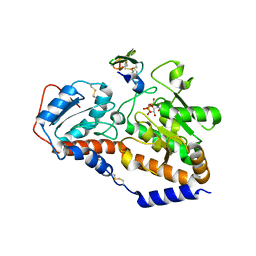 | | human POGLUT1 in complex with human Notch1 EGF12 S458T mutant and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurogenic locus notch homolog protein 1, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-12-20 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5T0A
 
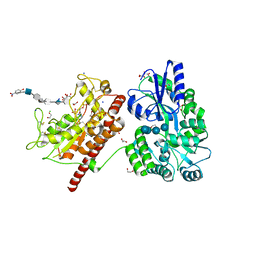 | | Crystal Structure of Heparan Sulfate 6-O-Sulfotransferase with bound PAP and heptasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5T05
 
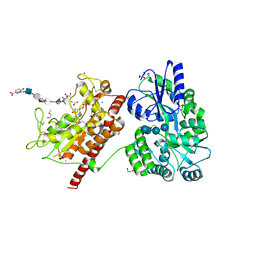 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and IdoA2S containing hexasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5TJ2
 
 | |
5T6R
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
5TTD
 
 | | Minor pilin FctB from S. pyogenes with engineered intramolecular isopeptide bond | | Descriptor: | FORMIC ACID, Maltose-binding periplasmic protein,Pilin isopeptide linkage domain protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Young, P.G, Kwon, H, Squire, C.J, Baker, E.N. | | Deposit date: | 2016-11-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Lys-Asn isopeptide bond into an immunoglobulin-like protein domain enhances its stability.
Sci Rep, 7, 2017
|
|
5T03
 
 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and glucuronic acid containing hexasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5VAW
 
 | |
5TJ4
 
 | |
