4E3O
 
 | |
4E0N
 
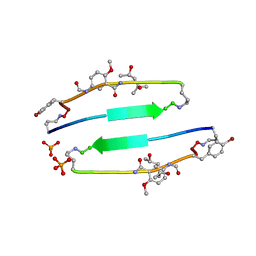 | | SVQIVYK segment from human Tau (305-311) displayed on 54-membered macrocycle scaffold (form II) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide SVQIVYK(ORN)EF(HAO)(4BF)K(ORN), PHOSPHATE ION | | Authors: | Zhao, M, Liu, C, Michael, S.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DET
 
 | |
4DGM
 
 | | Crystal Structure of maize CK2 in complex with the inhibitor apigenin | | Descriptor: | 1,2-ETHANEDIOL, 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Casein kinase II subunit alpha, ... | | Authors: | Lolli, G, Mazzorana, M, Battistutta, R. | | Deposit date: | 2012-01-26 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of protein kinase CK2 by flavonoids and tyrphostins. A structural insight.
Biochemistry, 51, 2012
|
|
5JLB
 
 | |
4X47
 
 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Anhydrosialidase, PHOSPHATE ION | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
5JJY
 
 | | Crystal structure of SETD2 bound to histone H3.3 K36M peptide | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yang, S, Zheng, X, Li, H. | | Deposit date: | 2016-04-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase
Genes Dev., 30, 2016
|
|
2AO6
 
 | | Crystal structure of the human androgen receptor ligand binding domain bound with TIF2(iii) 740-753 peptide and R1881 | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, 14-mer fragment of Nuclear receptor coactivator 2, androgen receptor | | Authors: | He, B, Gampe Jr, R.T, Kole, A.J, Hnat, A.T, Stanley, T.B, An, G, Stewart, E.L, Kalman, R.I, Minges, J.T, Wilson, E.M. | | Deposit date: | 2005-08-12 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance
Mol.Cell, 16, 2004
|
|
4DCD
 
 | | 1.6A resolution structure of PolioVirus 3C Protease Containing a covalently bound dipeptidyl inhibitor | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Genome polyprotein, ... | | Authors: | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.-C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | Deposit date: | 2012-01-17 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
4DEU
 
 | |
4X3G
 
 | | Crystal structure of SIAH1 SINA domain in complex with a USP19 peptide | | Descriptor: | E3 ubiquitin-protein ligase SIAH1, Ubiquitin carboxyl-terminal hydrolase 19, ZINC ION | | Authors: | Walker, J.R, Dong, A, Zhang, Q, Huang, X, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-11-28 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of SIAH1 SINA domain in complex with a USP19 peptide
To be published
|
|
4CAO
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(2-(3-(3-Fluorophenyl(propylamino)ethyl))quinolin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[2-[3-(3-fluorophenyl)propylamino]ethyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-10-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Simplified 2-Aminoquinoline-Based Scaffold for Potent and Selective Neuronal Nitric Oxide Synthase Inhibition.
J.Med.Chem., 57, 2014
|
|
4CAP
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(2-(3-(3-Fluorophenyl(propylamino)methyl))quinolin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-({[3-(3-fluorophenyl)propyl]amino}methyl)quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-10-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Simplified 2-Aminoquinoline-Based Scaffold for Potent and Selective Neuronal Nitric Oxide Synthase Inhibition.
J.Med.Chem., 57, 2014
|
|
4CAQ
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-((3-Chlorophenethylamino)ethyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[2-[2-(3-chlorophenyl)ethylamino]ethyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-10-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Simplified 2-Aminoquinoline-Based Scaffold for Potent and Selective Neuronal Nitric Oxide Synthase Inhibition.
J.Med.Chem., 57, 2014
|
|
8S4G
 
 | | Cryo-EM structure of the Anaphase-promoting complex/cyclosome (APC/C) bound to co-activator Cdh1 at 3.2 Angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2024-02-21 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | New structural features of the APC/C revealed by high resolution cryo-EM structures of apo-APC/C and the APC/C-CDH1-EMI1 complex
To Be Published
|
|
4CSI
 
 | | Crystal structure of the thermostable Cellobiohydrolase Cel7A from the fungus Humicola grisea var. thermoidea. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULASE, DI(HYDROXYETHYL)ETHER | | Authors: | Haddad-Momeni, M, Goedegebuur, F, Hansson, H, Karkehabadi, S, Askarieh, G, Mitchinson, C, Larenas, E, Stahlberg, J, Sandgren, M. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expression, Crystal Structure and Cellulase Activity of the Thermostable Cellobiohydrolase Cel7A from the Fungus Humicola Grisea Var. Thermoidea.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CFT
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 7-((3-Fluorophenethylamino)ethyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[2-[2-(3-fluorophenyl)ethylamino]ethyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Simplified 2-Aminoquinoline-Based Scaffold for Potent and Selective Neuronal Nitric Oxide Synthase Inhibition.
J.Med.Chem., 57, 2014
|
|
4FDL
 
 | | Crystal structure of Caspase-7 | | Descriptor: | Caspase-7 | | Authors: | Kabaleeswaran, V. | | Deposit date: | 2012-05-28 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | A class of allosteric caspase inhibitors identified by high-throughput screening.
Mol.Cell, 47, 2012
|
|
5L71
 
 | | Crystal structure of mouse phospholipid hydroperoxide glutathione peroxidase 4 (GPx4) | | Descriptor: | 1,2-ETHANEDIOL, Phospholipid hydroperoxide glutathione peroxidase, mitochondrial | | Authors: | Janowski, R, Scanu, S, Madl, T, Niessing, D. | | Deposit date: | 2016-06-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and solution structural studies of mouse phospholipid hydroperoxide glutathione peroxidase 4.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4FR3
 
 | | Crystal structure of human 14-3-3 sigma in complex with TASK-3 peptide and stabilizer 16-O-Me-FC-H | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(methoxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-glucopyranoside, 14-3-3 protein sigma, MAGNESIUM ION, ... | | Authors: | Ottmann, C, Anders, C, Schumacher, B. | | Deposit date: | 2012-06-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of k(+) channels at the cell surface.
Chem.Biol., 20, 2013
|
|
4CP8
 
 | | Structure of the amidase domain of allophanate hydrolase from Pseudomonas sp strain ADP | | Descriptor: | ALLOPHANATE HYDROLASE, MALONATE ION | | Authors: | Balotra, S, Newman, J, French, N, French, L, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-03 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of the Amidase Domain of Atzf, the Allophanate Hydrolase from the Cyanuric Acid-Mineralizing Multienzyme Complex.
Appl.Environ.Microbiol., 81, 2015
|
|
4D7M
 
 | | TetR(D) in complex with anhydrotetracycline and magnesium | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Werten, S, Dalm, D, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-11-25 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tetracycline Repressor Allostery Does not Depend on Divalent Metal Recognition.
Biochemistry, 53, 2014
|
|
5DOT
 
 | | Crystal Structure of Human Carbamoyl phosphate synthetase I (CPS1), apo form | | Descriptor: | 1,2-ETHANEDIOL, Carbamoyl-phosphate synthase [ammonia], mitochondrial, ... | | Authors: | Polo, L.M, de Cima, S, Fita, I, Rubio, V. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis.
Sci Rep, 5, 2015
|
|
5DOU
 
 | | Crystal Structure of Human Carbamoyl phosphate synthetase I (CPS1), ligand-bound form | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | de Cima, S, Polo, L.M, Fita, I, Rubio, V. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis.
Sci Rep, 5, 2015
|
|
4F6Z
 
 | | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-b-lactamase active site | | Descriptor: | Beta-lactamase, CITRATE ANION, ZINC ION | | Authors: | Horton, L.B, Shanker, S, Sankaran, B, Mikulski, R, Brown, N.G, Phillips, K, Lykissa, E, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2012-05-15 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site
Antimicrob.Agents Chemother., 56, 2012
|
|
