4B6U
 
 | | Solution structure of eIF4E3 in complex with m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E TYPE 3 | | Authors: | Osborne, M.J, Volpon, L, Kornblatt, J.A, Culkjovic-Kraljcic, B, Baguet, A, Borden, K.L.B. | | Deposit date: | 2012-08-15 | | Release date: | 2013-03-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Eif4E3 Acts as a Tumor Suppressor by Utilizing an Atypical Mode of Methyl-7-Guanosine CAP Recognition
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5L6K
 
 | | Crystal Structure of Human Carbonic Anhydrase II in Complex with a Quinoline Oligoamide Foldamer | | Descriptor: | 8-azanyl-4-(2-hydroxy-2-oxoethyloxy)quinoline-2-carboxylic acid, 8-azanyl-4-(2-methylpropoxy)quinoline-2-carboxylic acid, 8-azanyl-4-(3-azanylpropoxy)quinoline-2-carboxylic acid, ... | | Authors: | Jewginski, M, Langlois d'Estaintot, B, Granier, T, Huc, Y. | | Deposit date: | 2016-05-30 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
4AZR
 
 | | Human epidermal fatty acid-binding protein (FABP5) in complex with the endocannabinoid anandamide | | Descriptor: | CHLORIDE ION, FATTY ACID-BINDING PROTEIN, EPIDERMAL, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5WLE
 
 | |
5WLF
 
 | |
7M7E
 
 | | 6-Deoxyerythronolide B synthase (DEBS) hybrid module (M3/1) in complex with antibody fragment 1B2 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), 6-deoxyerythronolide-B synthase EryA2, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
4AWD
 
 | | Crystal structure of the beta-porphyranase BpGH16B (BACPLE_01689) from the human gut bacterium Bacteroides plebeius | | Descriptor: | BETA-PORPHYRANASE, CALCIUM ION, GLYCEROL | | Authors: | Hehemann, J.H, Kelly, A.G, Pudlo, N.A, Martens, E.C, Boraston, A.B. | | Deposit date: | 2012-06-01 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
4KTN
 
 | | Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor ((3S)-1-[2-(PYRIDO[2,3-B]PYRAZIN-7-YLSULFANYL)-9H-PYRIMIDO[4,5-B]INDOL-4-YL]PYRROLIDIN-3-AMINE) | | Descriptor: | (3S)-1-[2-(pyrido[2,3-b]pyrazin-7-ylsulfanyl)-9H-pyrimido[4,5-b]indol-4-yl]pyrrolidin-3-amine, DNA gyrase subunit B | | Authors: | Bensen, D.C, Akers-rodriguez, S, Tari, L.W. | | Deposit date: | 2013-05-20 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A new class of type iia topoisomerase inhibitors with
broad-spectrum antibacterial activity
To be Published
|
|
6QNZ
 
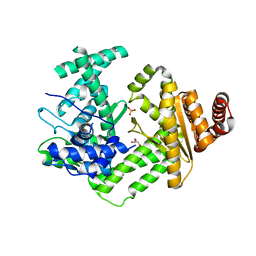 | | Crystal structure of the site-specific DNA nickase N.BspD6I E418A Mutant | | Descriptor: | GLYCEROL, Heterodimeric restriction endonuclease R.BspD6I large subunit, PHOSPHATE ION | | Authors: | Artyukh, R.I, Kachalova, G.S, Yunusova, A.K, Gabdulkhakov, A.G, Fatkhullin, B.F, Atanasov, B.P, Perevyazova, T.A, Popov, A.N, Zheleznaya, L.A. | | Deposit date: | 2019-02-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The key role of E418 carboxyl group in the formation of Nt.BspD6I nickase active site: Structural and functional properties of Nt.BspD6I E418A mutant.
J.Struct.Biol., 210, 2020
|
|
7MRZ
 
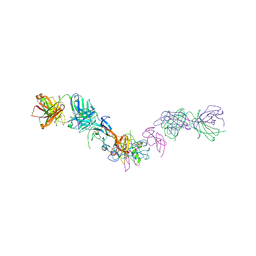 | | Structure of GDF11 bound to fused ActRIIB-ECD and Alk4-ECD with Anti-ActRIIB Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B,Activin receptor type-1B, Fab Heavy Chain, ... | | Authors: | Goebel, E.J, Kattamuri, C, Gipson, G.R, Thompson, T.B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
5WCU
 
 | |
5W7D
 
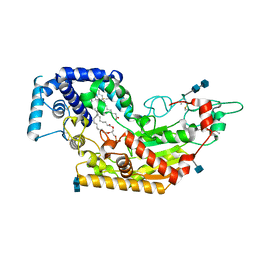 | | Murine acyloxyacyl hydrolase (AOAH), S262A mutant | | Descriptor: | 1,2-DISTEAROYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the mammalian lipopolysaccharide detoxifier.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6WJB
 
 | | UDP-GlcNAc C4-epimerase from Pseudomonas protegens in complex with NAD and UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Pfoh, R, Robinson, H, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
8BK6
 
 | | A truncated structure of LpMIP with bound inhibitor JK095. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase | | Authors: | Whittaker, J.J, Guskov, A, Hellmich, A.U, Goretzki, B. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
5L9E
 
 | | CRYSTAL STRUCTURE OF HUMAN CARBONIC ANHYDRASE II IN COMPLEX WITH A QUINOLINE OLIGOAMIDE FOLDAMER | | Descriptor: | 8-azanyl-4-(2-hydroxy-2-oxoethyloxy)quinoline-2-carboxylic acid, 8-azanyl-4-(2-methylpropoxy)quinoline-2-carboxylic acid, Carbonic anhydrase 2, ... | | Authors: | Vallade, M, Langlois d'Estaintot, B, Granier, T, Huc, I. | | Deposit date: | 2016-06-10 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CRYSTAL STRUCTURE OF HUMAN CARBONIC ANHYDRASE II IN COMPLEX WITH A QUINOLINE OLIGOAMIDE FOLDAMER
To Be Published
|
|
3MEU
 
 | | Crystal structure of SGF29 in complex with H3R2me2sK4me3 | | Descriptor: | Histone H3, SAGA-associated factor 29 homolog, SULFATE ION | | Authors: | Bian, C.B, Xu, C, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
4B92
 
 | |
6X5B
 
 | | Symmetric model of CD4- and 17-bound B41 HIV-1 Env SOSIP in complex with small molecule GO52 | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2020-05-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
8BO1
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AZIDE ION, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
1RJW
 
 | |
8BR1
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-22 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
6WQP
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, Endoglucanase, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
5A7O
 
 | | Crystal structure of human JMJD2A in complex with compound 42 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-(2-methoxyethanoylamino)-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
4BFQ
 
 | | Assembly of a triple pi-stack of ligands in the binding site of Aplysia californica acetylcholine binding protein (AChBP) | | Descriptor: | 4,6-dimethyl-N'-(3-pyridin-2-ylisoquinolin-1-yl)pyrimidine-2-carboximidamide, GLYCEROL, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Stornaiuolo, M, De Kloe, G.E, Rucktooa, P, Fish, A, van Elk, R, Edink, E.S, Bertrand, D, Smit, A.B, de Esch, I.J.P, Sixma, T.K. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Assembly of a Pi-Pi Stack of Ligands in the Binding Site of an Acetylcholine Binding Protein
Nat.Commun., 4, 2013
|
|
5WPJ
 
 | |
