8R1W
 
 | | Pim1 in complex with 4-(5-(4-aminophenyl)-2H-1,2,3-triazol-4-yl)benzoic acid and Pimtide | | Descriptor: | 4-(5-(4-aminophenyl)-2H-1,2,3-triazol-4-yl)benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-11-02 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pim1 in complex with 4-(5-(4-aminophenyl)-2H-1,2,3-triazol-4-yl)benzoic acid and Pimtide
To Be Published
|
|
8R27
 
 | |
7LQD
 
 | | Structure of Human MPS1 (TTK) covalently bound to RMS-07 inhibitor | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, Dual specificity protein kinase TTK or monopolar spindle 1 kinase, GLYCEROL, ... | | Authors: | Santiago, A.S, dos Reis, C.V, Serafim, R.A.M, Massirer, K.B, Arruda, P, Edwards, A.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-02-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of the First Covalent Monopolar Spindle Kinase 1 (MPS1/TTK) Inhibitor.
J.Med.Chem., 65, 2022
|
|
5J87
 
 | |
9DR5
 
 | |
7QRS
 
 | | Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 | | Descriptor: | ACETATE ION, GLYCEROL, Protein Tax-1, ... | | Authors: | Javorsky, A, Soares da Costa, T.P, Mackie, E.R, Humbert, P.O, Kvansakul, M. | | Deposit date: | 2022-01-12 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 PBM.
Febs J., 290, 2023
|
|
6SSN
 
 | | RNASE 3/1 version3 | | Descriptor: | GLYCEROL, PHOSPHATE ION, RNase 3/1 version3 | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2019-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Exploring the RNase A scaffold to combine catalytic and antimicrobial activities. Structural characterization of RNase 3/1 chimeras.
Front Mol Biosci, 9, 2022
|
|
7LE0
 
 | | HIV-1 Protease WT (NL4-3) in Complex with a Darunavir Derivative | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(1,3-benzothiazol-6-yl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
7LE2
 
 | | HIV-1 Protease WT (NL4-3) in Complex with UMass4 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{(1,3-benzodioxol-5-ylsulfonyl)[(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
7TZC
 
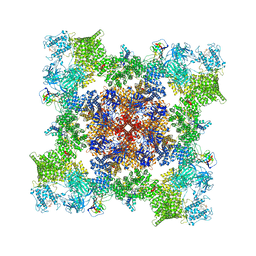 | | A drug and ATP binding site in type 1 ryanodine receptor | | Descriptor: | (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Melville, Z, Dridi, H, Yuan, Q, Reiken, S, Anetta, W, Liu, Y, Clarke, O.B, Marks, A.R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | A drug and ATP binding site in type 1 ryanodine receptor.
Structure, 30, 2022
|
|
7B5A
 
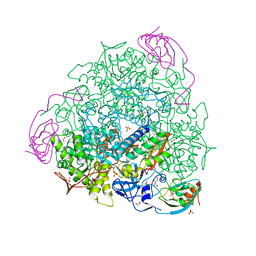 | | X-ray crystal structure of Sporosarcina pasteurii urease inhibited by Ag(PEt3)2NO3 determined at 1.97 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2020-12-03 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Kinetic and structural analysis of the inactivation of urease by mixed-ligand phosphine halide Ag(I) complexes.
J.Inorg.Biochem., 218, 2021
|
|
7B58
 
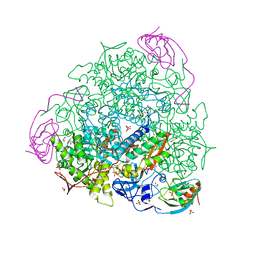 | | X-ray crystal structure of Sporosarcina pasteurii urease inhibited by Ag(PEt3)Cl determined at 1.72 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2020-12-03 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetic and structural analysis of the inactivation of urease by mixed-ligand phosphine halide Ag(I) complexes.
J.Inorg.Biochem., 218, 2021
|
|
7B59
 
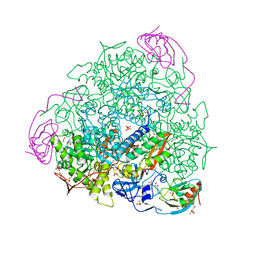 | | X-ray crystal structure of Sporosarcina pasteurii urease inhibited by Ag(PEt3)Br determined at 1.63 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2020-12-03 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Kinetic and structural analysis of the inactivation of urease by mixed-ligand phosphine halide Ag(I) complexes.
J.Inorg.Biochem., 218, 2021
|
|
5MWM
 
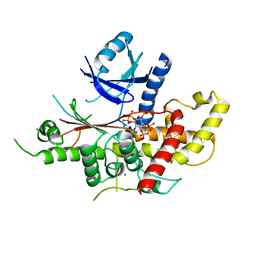 | | INOSITOL 1,3,4,5,6-PENTAKISPHOSPHATE 2-KINASE FROM M. MUSCULUS IN COMPLEX WITH IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol-pentakisphosphate 2-kinase, ZINC ION | | Authors: | Franco-Echevarria, E, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of mammalian inositol 1,3,4,5,6-pentakisphosphate 2-kinase reveals a new zinc-binding site and key features for protein function.
J. Biol. Chem., 292, 2017
|
|
8AKP
 
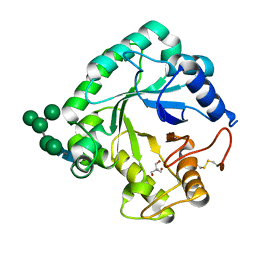 | | Crystal structure of the catalytic domain of G7048 from Penicillium sumatraense | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, catalytic domain of G7048 | | Authors: | Troilo, F, Scafati, V, Giovannoni, M, Mattei, B, Benedetti, M, Angelucci, F, Di Matteo, A. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of two 1,3-beta-glucan-modifying enzymes from Penicillium sumatraense reveals new insights into 1,3-beta-glucan metabolism of fungal saprotrophs.
Biotechnol Biofuels Bioprod, 15, 2022
|
|
5MF4
 
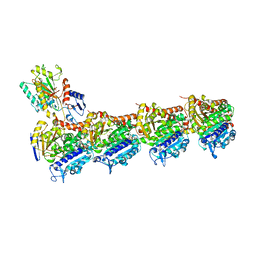 | | Tubulin-Dictyostatin complex | | Descriptor: | (3~{Z},5~{E},7~{R},8~{S},10~{S},11~{Z},13~{S},14~{R},15~{S},17~{S},20~{R},21~{S},22~{S})-22-[(2~{S},3~{Z})-hexa-3,5-dien-2-yl]-7,13,15,17,21-pentamethyl-8,10,14,20-tetrakis(oxidanyl)-1-oxacyclodocosa-3,5,11-trien-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Trigili, C, Barasoain, I, Sanchez-Murcia, P.A, Bargsten, K, Redondo-Horcajo, M, Nogales, A, Gardner, N.M, Meyer, A, Naylor, G.J, Gomez-Rubio, E, Gago, F, Steinmetz, M.O, Paterson, I, Prota, A.E, Diaz, J.F. | | Deposit date: | 2016-11-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determinants of the Dictyostatin Chemotype for Tubulin Binding Affinity and Antitumor Activity Against Taxane- and Epothilone-Resistant Cancer Cells.
ACS Omega, 1, 2016
|
|
7MOS
 
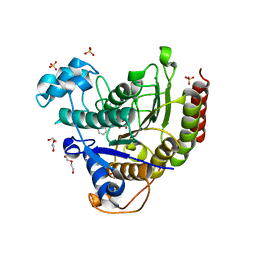 | | Structure of HDAC2 in complex with a macrocyclic inhibitor (compound 4) | | Descriptor: | (3S,18S,20aR)-18-(6,6-dihydroxyoctyl)-1,5,6,7,8,18,19,20a-octahydro-4H-14,17-epiminoazeto[1,2-g][1,7,10,13]benzoxatriazacycloheptadecin-20(2H)-one, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
6W5Y
 
 | | Inferred receptor binding domain of human endogenous retrovirus envelope EnvP(b)1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, EnvP(b)1 inferred receptor binding domain, ... | | Authors: | McCarthy, K.R. | | Deposit date: | 2020-03-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Receptor Binding Domain of EnvP(b)1, an Endogenous Retroviral Envelope Protein Expressed in Human Tissues.
Mbio, 11, 2020
|
|
9FN8
 
 | | Crystal structure of human carboanhydrase XII with 4-benzyl-5,7,8-trifluoro-3,4-dihydro-2H-benzo[b][1,4]thiazine-6-sulfonamide 1,1-dioxide | | Descriptor: | 1,2-ETHANEDIOL, 5,7,8-tris(fluoranyl)-1,1-bis(oxidanylidene)-4-(phenylmethyl)-2,3-dihydro-1$l^{6},4-benzothiazine-6-sulfonamide, Carbonic anhydrase 12, ... | | Authors: | Manakova, E, Paketuryte, V, Trumpickaite, G, Smirnov, A, Vaskevicius, A. | | Deposit date: | 2024-06-09 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Design of Rigid Compounds to Enhance Selectivity for Carbonic Anhydrase IX.
Chemistry, 31, 2025
|
|
9FN7
 
 | | Crystal structure of human carboanhydrase XII with 5,7,8-trifluoro-4-(3-propylhexyl)-3,4-dihydro-2H-benzo[b][1,4]thiazine-6-sulfonamide 1,1-dioxide | | Descriptor: | 1,2-ETHANEDIOL, 5,7,8-tris(fluoranyl)-1,1-bis(oxidanylidene)-4-(3-propylhexyl)-2,3-dihydro-1$l^{6},4-benzothiazine-6-sulfonamide, Carbonic anhydrase 12, ... | | Authors: | Manakova, E, Paketuryte, V, Trumpickaite, G, Smirnov, A, Vaskevicius, A. | | Deposit date: | 2024-06-09 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Design of Rigid Compounds to Enhance Selectivity for Carbonic Anhydrase IX.
Chemistry, 31, 2025
|
|
5MXM
 
 | | The X-ray structure of human M190I phosphoglycerate kinase 1 mutant | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ilari, A, Fiorillo, A, Cipollone, A, Petrosino, M. | | Deposit date: | 2017-01-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
7BTV
 
 | | Crystal structure of EHMT2 SET domain in complex with compound 5. | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N~2~-{4-methoxy-3-[3-(pyrrolidin-1-yl)propoxy]phenyl}-N~4~,6-dimethylpyrimidine-2,4-diamine, S-ADENOSYLMETHIONINE, ... | | Authors: | Suzuki, M, Mizuno, T, Katayama, K. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel histone lysine methyltransferase G9a/GLP (EHMT2/1) inhibitors: Design, synthesis, and structure-activity relationships of 2,4-diamino-6-methylpyrimidines.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5MAC
 
 | | Crystal structure of decameric Methanococcoides burtonii Rubisco complexed with 2-carboxyarabinitol bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Gunn, L.H, Valegard, K, Andersson, I. | | Deposit date: | 2016-11-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique structural domain in Methanococcoides burtonii ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) acts as a small subunit mimic.
J. Biol. Chem., 292, 2017
|
|
6QLB
 
 | | Calpain small subunit 1, RNA-binding protein Hfq | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, CALCIUM ION, ... | | Authors: | Rizkallah, P.J, Cresser-Brown, J.O. | | Deposit date: | 2019-01-31 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Calpain small subunit 1, RNA-binding protein Hfq
Acta Crystallogr.,Sect.F, 2020
|
|
8R7M
 
 | | CTX-M14 in complex with boric acid and 1,2-diol boric ester | | Descriptor: | BORIC ACID, Beta-lactamase, GLYCEROL, ... | | Authors: | Werner, N, Prester, A, Hinrichs, W, Perbandt, M, Betzel, C. | | Deposit date: | 2023-11-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
