7P7R
 
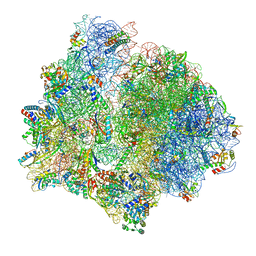 | | PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state I | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
9O9N
 
 | | Crystal structure of PPARgamma ligand binding domain (LBD) in complex with NCoR1 corepressor peptide and inverse agonist FX-909 | | Descriptor: | (3P)-3-(5,7-difluoro-4-oxo-1,4-dihydroquinolin-2-yl)-4-(methanesulfonyl)benzonitrile, Nuclear receptor corepressor 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Laughlin, Z.T, Dong, J, Harp, J.M, Kojetin, D.J. | | Deposit date: | 2025-04-18 | | Release date: | 2025-05-14 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of PPAR gamma-Mediated Transcriptional Repression by the Covalent Inverse Agonist FX-909.
J.Med.Chem., 68, 2025
|
|
7MIT
 
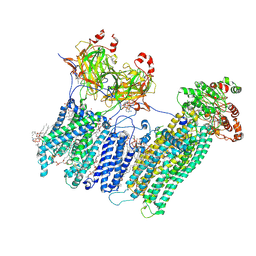 | | Vascular KATP channel: Kir6.1 SUR2B propeller-like conformation 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ... | | Authors: | Sung, M.W, Shyng, S.L. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Vascular K ATP channel structural dynamics reveal regulatory mechanism by Mg-nucleotides.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6OUD
 
 | |
8G74
 
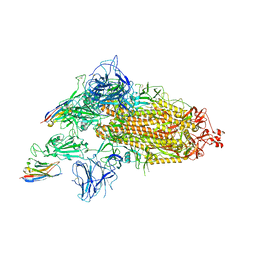 | | SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
7PC8
 
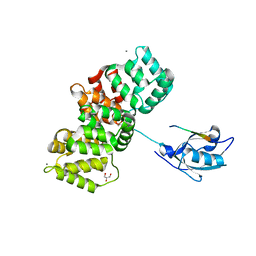 | | The PDZ domain of SNTG1 complexed with the phosphomimetic mutant PDZ-binding motif of RSK1 | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-1-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
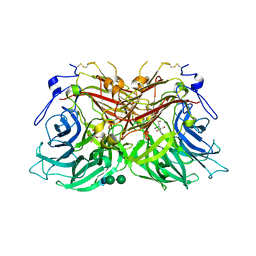
8BES
 
 | |
6X7C
 
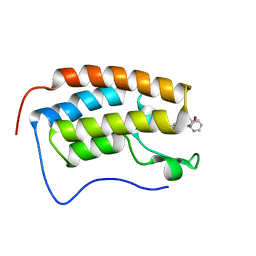 | | BRD4 Bromodomain 1 in complex with multi-action inhibitor SRX3212 | | Descriptor: | 7-[5-(morpholin-4-yl)-7-oxo-7H-thieno[3,2-b]pyran-3-yl]-N-[(pyridin-3-yl)methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxamide, Bromodomain-containing protein 4 | | Authors: | Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-29 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of thienopyranone-based BET inhibitors that bind multiple synthetic lethality targets.
Sci Rep, 10, 2020
|
|
6XMX
 
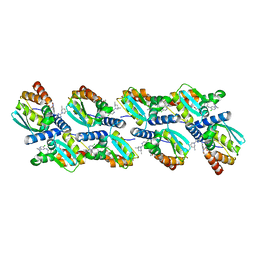 | | Cryo-EM structure of BCL6 bound to BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Yoon, H, Burman, S.S.R, Hunkeler, M, Nowak, R.P, Fischer, E.S. | | Deposit date: | 2020-07-01 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Small-molecule-induced polymerization triggers degradation of BCL6.
Nature, 588, 2020
|
|
8GES
 
 | | R. hominis 2 beta-glucuronidase bound to UNC10201652-glucuronide | | Descriptor: | 8-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5-(morpholin-4-yl)-1,2,3,4-tetrahydro[1,2,3]triazino[4',5':4,5]thieno[2,3 -c]isoquinoline, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
6OPH
 
 | | phosphorylated ERK2 with GDC-0994 | | Descriptor: | 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Mitogen-activated protein kinase 1 | | Authors: | Vigers, G.P, Smith, D. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation loop dynamics are controlled by conformation-selective inhibitors of ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O7Q
 
 | |
8GA2
 
 | | Bromodomain of CBP liganded with inhibitor iCBP5 | | Descriptor: | (6S)-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}-1-phenylpiperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
6X7D
 
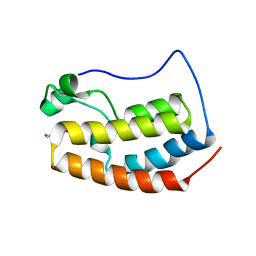 | |
6X7P
 
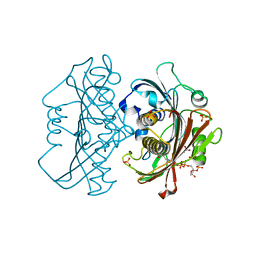 | | LnmK in complex with 2-sulfonate-propionyl-oxa(dethia)-CoA | | Descriptor: | (2~{R})-1-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethoxy]-1-oxidanylidene-propane-2-sulfonic acid, (2~{S})-1-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethoxy]-1-oxidanylidene-propane-2-sulfonic acid, Conserved hypthetical protein, ... | | Authors: | Stunkard, L.M, Kick, B.J, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of LnmK, a Bifunctional Acyltransferase/Decarboxylase, with Substrate Analogues Reveal the Basis for Selectivity and Stereospecificity.
Biochemistry, 60, 2021
|
|
6X6H
 
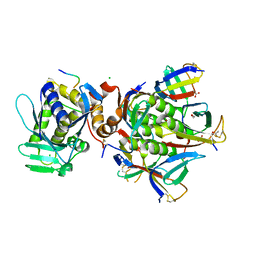 | |
7NBM
 
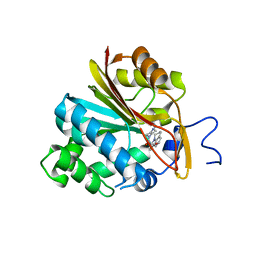 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with the bisubstrate-like inhibitor (33) | | Descriptor: | (E)-3-((5,6-dihydro-2H,4H-thiazolo[5,4,3-ij]quinolin-2-ylidene)amino)-2-hydroxy-1-(4-(isoquinolin-5-yl)piperazin-1-yl)-2-methylpropan-1-one, Nicotinamide N-methyltransferase | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2021-01-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Novel Inhibitors of Nicotinamide- N -Methyltransferase for the Treatment of Metabolic Disorders.
Molecules, 26, 2021
|
|
7NDG
 
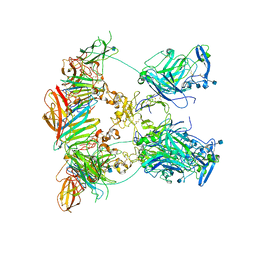 | | Cryo-EM structure of the ternary complex between Netrin-1, Neogenin and Repulsive Guidance Molecule B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neogenin, ... | | Authors: | Robinson, R.A, Griffiths, S.C, van de Haar, L.L, Malinauskas, T, van Battum, E.Y, Zelina, P, Schwab, R.A, Karia, D, Malinauskaite, L, Brignani, S, van den Munkhof, M, Dudukcu, O, De Ruiter, A.A, Van den Heuvel, D.M.A, Bishop, B, Elegheert, J, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2021-02-01 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.98 Å) | | Cite: | Simultaneous binding of Guidance Cues NET1 and RGM blocks extracellular NEO1 signaling.
Cell, 184, 2021
|
|
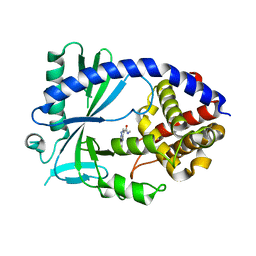
6NFG
 
 | |
9OF2
 
 | | Dimer of HIF-2a-ARNT Heterodimers Complexed on 51-bp HRE/HAS | | Descriptor: | 51-nt Hypoxia Response Element (Forward), 51-nt Hypoxia Response Element (Reverse), Aryl hydrocarbon receptor nuclear translocator, ... | | Authors: | Closson, J.D, Xu, X, Gardner, K.H. | | Deposit date: | 2025-04-29 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Context-Dependent Variability Of HIF Heterodimers Influences Interactions With Macromolecular And Small Molecule Partners.
Biorxiv, 2025
|
|
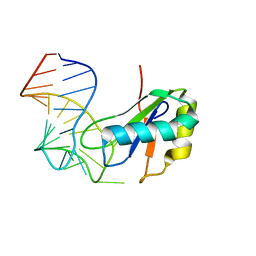
6XH3
 
 | |
9OFV
 
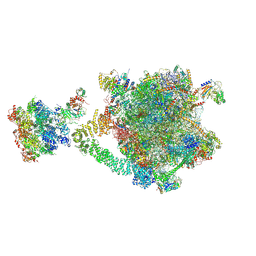 | | Consensus reconstruction of the eukaryotic Ribosome-associated Quality Control complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Li, W, Cahoon, T, Shen, P.S. | | Deposit date: | 2025-04-30 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Mechanism of nascent chain removal by the ribosome-associated quality control complex.
Nat Commun, 16, 2025
|
|
6XH2
 
 | |
6OL9
 
 | | Structure of the M5 muscarinic acetylcholine receptor (M5-T4L) bound to tiotropium | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Vuckovic, Z, Christopoulos, A, Thal, D.M. | | Deposit date: | 2019-04-16 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Crystal structure of the M5muscarinic acetylcholine receptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6XN2
 
 | | Crystal structure of the GH43_1 enzyme from Xanthomonas citri complexed with xylotriose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | Deposit date: | 2020-07-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
