1A8L
 
 | | PROTEIN DISULFIDE OXIDOREDUCTASE FROM ARCHAEON PYROCOCCUS FURIOSUS | | Descriptor: | PROTEIN DISULFIDE OXIDOREDUCTASE, ZINC ION | | Authors: | Ren, B, Tibbelin, G, Pascale, D, Rossi, M, Bartolucci, S, Ladenstein, R. | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus contains two thioredoxin fold units.
Nat.Struct.Biol., 5, 1998
|
|
7AK9
 
 | | Structure of Salmonella TacT3 toxin bound to TacA3 antitoxin C-terminal peptide | | Descriptor: | ABC transporter, Acetyltransferase, DEPHOSPHO COENZYME A | | Authors: | Grabe, G.J, Morgan, R.M.L, Hare, S.A, Helaine, S. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
7AK7
 
 | | Structure of Salmonella TacT2 toxin bound to TacA2 antitoxin | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, CHLORIDE ION, ... | | Authors: | Grabe, G.J, Morgan, R.M.L, Hare, S.A, Helaine, S. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
7AK8
 
 | | Structure of Salmonella TacT1 toxin bound to TacA1 antitoxin C-terminal peptide | | Descriptor: | ACETYL COENZYME *A, GCN5 family acetyltransferase, GLYCEROL, ... | | Authors: | Grabe, G.J, Morgan, R.M.L, Helaine, S, Hare, S.A. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
4X47
 
 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Anhydrosialidase, PHOSPHATE ION | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
4X6K
 
 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Siastatin B | | Descriptor: | (2S,3R,4S,5S)-2-(acetylamino)-5-carboxy-3,4-dihydroxypiperidinium, ACETYL GROUP, Anhydrosialidase | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
4X4A
 
 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with 2,7-Anhydro-Neu5Ac | | Descriptor: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, ACETYL GROUP, Anhydrosialidase, ... | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
8OO0
 
 | | Chaetomium thermophilum Methionine Aminopeptidase 2 autoproteolysis product at the 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S0, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
8ONZ
 
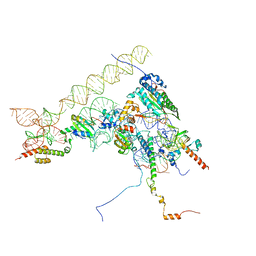 | | Chaetomium thermophilum Methionine Aminopeptidase 2 at the 80S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 60S ribosomal protein L25-like protein, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
8ONX
 
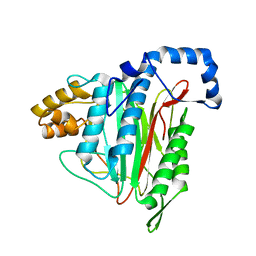 | | High resolution structure of Chaetomium thermophilum MAP2 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
8ONY
 
 | | Human Methionine Aminopeptidase 2 at the 80S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 60S ribosomal protein L19, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
6WN2
 
 | |
7KXS
 
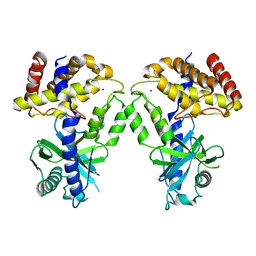 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2020-12-04 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7CM5
 
 | | Full-length Sarm1 in a self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
7CM7
 
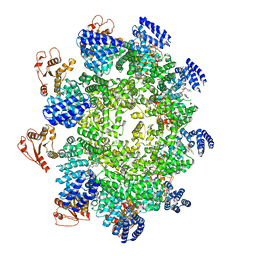 | | NAD+-bound Sarm1 E642A in the self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
7CM6
 
 | | NAD+-bound Sarm1 in the self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
7LZ3
 
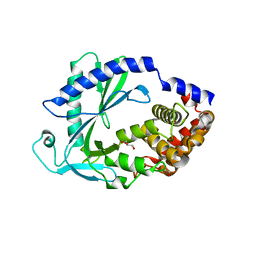 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, GLYCEROL, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CLS
 
 | |
5N4B
 
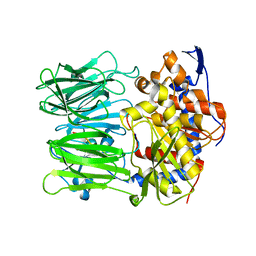 | | Prolyl oligopeptidase B from Galerina marginata bound to 25mer macrocyclization substrate - S577A mutant | | Descriptor: | Alpha-amanitin proprotein, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5N4E
 
 | | Prolyl oligopeptidase B from Galerina marginata bound to 35mer hydrolysis and macrocyclization substrate - H698A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
6MJW
 
 | | human cGAS catalytic domain bound with the inhibitor G150 | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Lama, L, Adura, C, Xie, W, Tomita, D, Kamei, T, Kuryavyi, V, Gogakos, T, Steinberg, J.I, Miller, M, Ramos-Espiritu, L, Asano, Y, Hashizume, S, Aida, J, Imaeda, T, Okamoto, R, Jennings, A.J, Michinom, M, Kuroita, T, Stamford, A, Gao, P, Meinke, P, Glickman, J.F, Patel, D.J, Tuschl, T. | | Deposit date: | 2018-09-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression.
Nat Commun, 10, 2019
|
|
6TDO
 
 | | Crystal structure of the disulfide engineered HLA-A0201 molecule in complex with one GM dipeptide in the A pocket. | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCINE, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-09 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
6TDQ
 
 | | Crystal structure of the disulfide engineered HLA-A0201 molecule in complex with one GM dipeptide in the A pocket and one GM dipeptide in the F pocket. | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
6TDS
 
 | | Crystal structure of the disulfide engineered HLA-A0201 molecule without peptide bound after NaCl wash | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
5N4C
 
 | | Prolyl oligopeptidase B from Galerina marginata bound to 35mer hydrolysis and macrocyclization substrate - S577A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
