4PTF
 
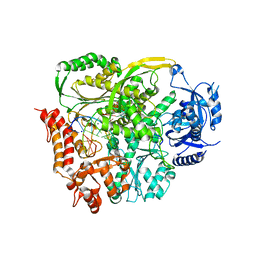 | | Ternary crystal structure of yeast DNA polymerase epsilon with template G | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', ... | | 著者 | Jain, R, Rajashankar, K.R, Buku, A, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | 登録日 | 2014-03-10 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.809 Å) | | 主引用文献 | Crystal Structure of Yeast DNA Polymerase epsilon Catalytic Domain.
Plos One, 9, 2014
|
|
8OHM
 
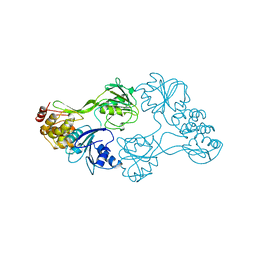 | |
1XSG
 
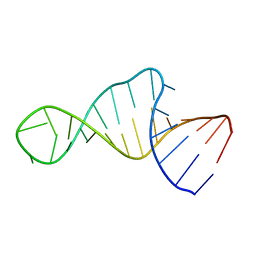 | |
1XSH
 
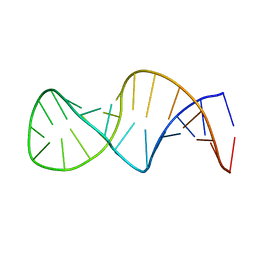 | |
280D
 
 | |
2LHP
 
 | |
2LUB
 
 | |
2JWV
 
 | |
2RVO
 
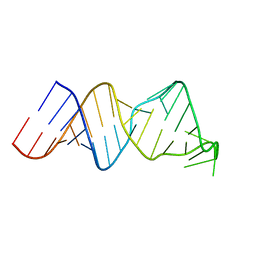 | | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish | | 分子名称: | RNA (34-MER) | | 著者 | Otsu, M, Norose, N, Arai, N, Terao, R, Kajikawa, M, Okada, N, Kawai, G. | | 登録日 | 2016-02-03 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish.
J. Biochem., 162, 2017
|
|
2L1F
 
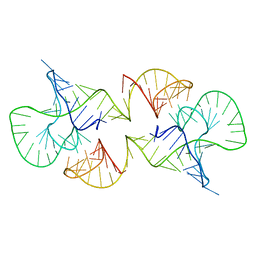 | | Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography | | 分子名称: | RNA (65-MER), RNA (66-MER) | | 著者 | Summers, M.F, Irobalieva, R.N, Tolbert, B, Smalls-Manty, A, Iyalla, K, Loeliger, K, D'Souza, V, Khant, H, Schmid, M, Garcia, E, Telesnitsky, A, Chiu, W, Miyazaki, Y. | | 登録日 | 2010-07-28 | | 公開日 | 2010-10-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography.
J.Mol.Biol., 404, 2010
|
|
3NKY
 
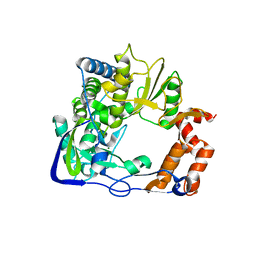 | | Structure of a mutant P44S of Foot-and-mouth disease Virus RNA-dependent RNA polymerase | | 分子名称: | 3D polymerase, MAGNESIUM ION | | 著者 | Agudo, R, Ferrer-Orta, C, Arias, A, Perez-Luque, R, Verdaguer, N, Domingo, E. | | 登録日 | 2010-06-21 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | A multi-step process of viral adaptation to a mutagenic nucleoside analogue by modulation of transition types leads to extinction-escape.
Plos Pathog., 6, 2010
|
|
4LT6
 
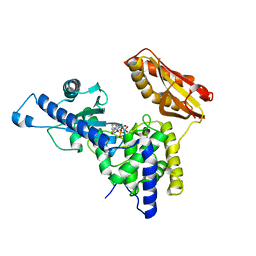 | | Crystal Structure of human poly(A) polymerase gamma | | 分子名称: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Poly(A) polymerase gamma | | 著者 | Yang, Q, Nausch, L, Martin, G, Keller, W, Doublie, S. | | 登録日 | 2013-07-23 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Crystal structure of human poly(a) polymerase gamma reveals a conserved catalytic core for canonical poly(a) polymerases.
J.Mol.Biol., 426, 2014
|
|
6KLE
 
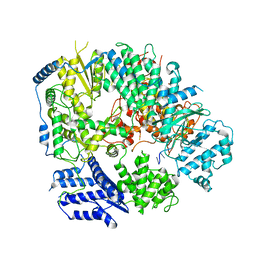 | | Monomeric structure of Machupo virus polymerase bound to vRNA promoter | | 分子名称: | MANGANESE (II) ION, RNA (5'-R(*GP*CP*CP*UP*AP*GP*GP*AP*UP*CP*CP*AP*CP*UP*GP*UP*GP*CP*G)-3'), RNA-directed RNA polymerase L, ... | | 著者 | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | 登録日 | 2019-07-30 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
6KLD
 
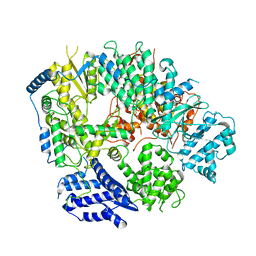 | | Structure of apo Machupo virus polymerase | | 分子名称: | MANGANESE (II) ION, RNA-directed RNA polymerase L, ZINC ION | | 著者 | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | 登録日 | 2019-07-30 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
6LBJ
 
 | |
6LBK
 
 | |
9F37
 
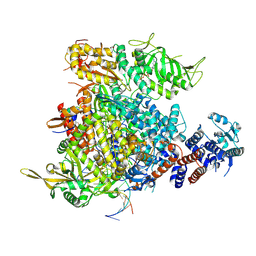 | |
1U76
 
 | |
9F2R
 
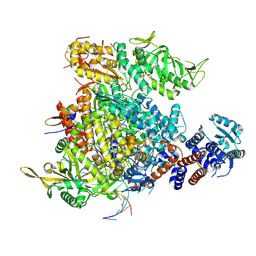 | |
6X7F
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B2 (TTC-B2) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-05-29 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X7K
 
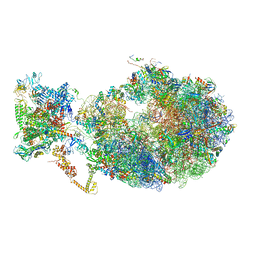 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-05-30 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XDR
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 27 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-06-11 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XDQ
 
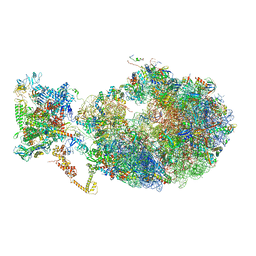 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 30 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-06-11 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XII
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 24 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-06-20 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XGF
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 30 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-06-17 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
