8DYT
 
 | | Cryo-EM structure of 227 Fab in complex with (NPNA)8 peptide | | Descriptor: | 227 Fab heavy chain, 227 Fab light chain, Circumsporozoite protein | | Authors: | Martin, G.M, Ward, A.B. | | Deposit date: | 2022-08-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Affinity-matured homotypic interactions induce spectrum of PfCSP structures that influence protection from malaria infection.
Nat Commun, 14, 2023
|
|
8DYX
 
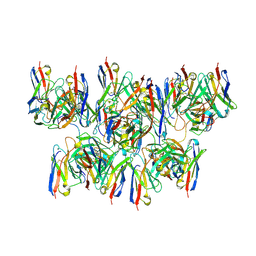 | |
8DZ4
 
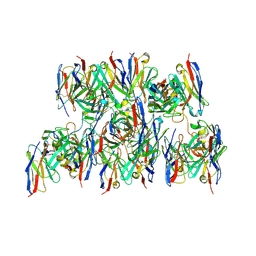 | |
8DZ3
 
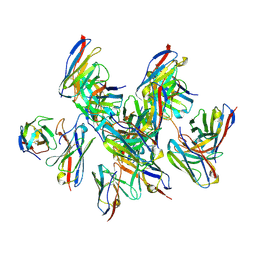 | |
8DYY
 
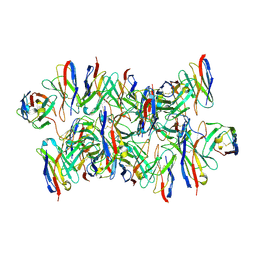 | |
8DZ5
 
 | |
8DYW
 
 | |
6BAA
 
 | | Cryo-EM structure of the pancreatic beta-cell KATP channel bound to ATP and glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Martin, G.M, Yoshioka, C, Shyng, S.L. | | Deposit date: | 2017-10-12 | | Release date: | 2017-11-01 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Anti-diabetic drug binding site in a mammalian KATPchannel revealed by Cryo-EM.
Elife, 6, 2017
|
|
1F5A
 
 | | CRYSTAL STRUCTURE OF MAMMALIAN POLY(A) POLYMERASE | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, POLY(A) POLYMERASE, ... | | Authors: | Martin, G, Keller, W, Doublie, S. | | Deposit date: | 2000-06-13 | | Release date: | 2000-09-13 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP.
EMBO J., 19, 2000
|
|
5TWV
 
 | | Cryo-EM structure of the pancreatic ATP-sensitive K+ channel SUR1/Kir6.2 in the presence of ATP and glibenclamide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ATP-sensitive inward rectifier potassium channel 11 | | Authors: | Martin, G.M, Yoshioka, C, Chen, J.Z, Shyng, S.L. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-25 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating.
Elife, 6, 2017
|
|
1Q79
 
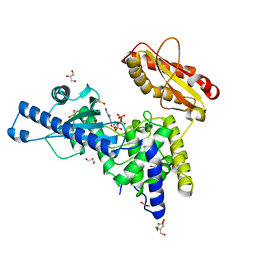 | | CRYSTAL STRUCTURE OF MAMMALIAN POLY(A) POLYMERASE | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Martin, G, Moglich, A, Keller, W, Doublie, S. | | Deposit date: | 2003-08-16 | | Release date: | 2004-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase.
J.Mol.Biol., 341, 2004
|
|
7LX2
 
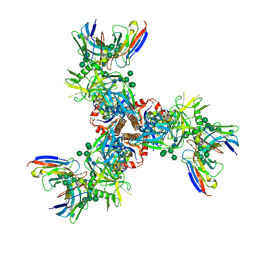 | | Cryo-EM structure of ConSOSL.UFO.664 (ConS) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Env glycoprotein gp160, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
7LX3
 
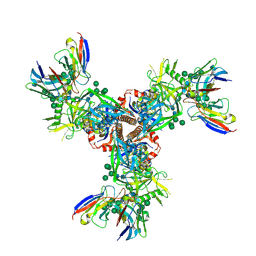 | | Cryo-EM structure of EDC-crosslinked ConSOSL.UFO.664 (ConS-EDC) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Env glycoprotein gp160, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
7LXM
 
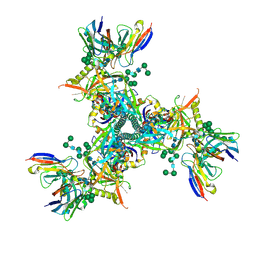 | | Cryo-EM structure of ConM SOSIP.v7 (ConM) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env glycoprotein gp120, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
7LXN
 
 | | Cryo-EM structure of EDC-crosslinked ConM SOSIP.v7 (ConM-EDC) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env glycoprotein gp120, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
1Q78
 
 | | Crystal structure of poly(A) polymerase in complex with 3'-dATP and magnesium chloride | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Poly(A) polymerase alpha | | Authors: | Martin, G, Moglich, A, Keller, W, Doublie, S. | | Deposit date: | 2003-08-16 | | Release date: | 2004-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase.
J.Mol.Biol., 341, 2004
|
|
8EH5
 
 | | Cryo-EM structure of L9 Fab in complex with rsCSP | | Descriptor: | Circumsporozoite protein, L9 Heavy chain, L9 Light chain | | Authors: | Martin, G.M, Ward, A.B. | | Deposit date: | 2022-09-13 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of epitope selectivity and potent protection from malaria by PfCSP antibody L9.
Nat Commun, 14, 2023
|
|
1LFD
 
 | | CRYSTAL STRUCTURE OF THE ACTIVE RAS PROTEIN COMPLEXED WITH THE RAS-INTERACTING DOMAIN OF RALGDS | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RALGDS, ... | | Authors: | Huang, L, Hofer, F, Martin, G.S, Kim, S.-H. | | Deposit date: | 1998-04-29 | | Release date: | 1999-05-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction of Ras with RalGDS.
Nat.Struct.Biol., 5, 1998
|
|
1LXD
 
 | | CRYSTAL STRUCTURE OF THE RAS INTERACTING DOMAIN OF RALGDS, A GUANINE NUCLEOTIDE DISSOCIATION STIMULATOR OF RAL PROTEIN | | Descriptor: | RALGDSB | | Authors: | Huang, L, Weng, X.W, Hofer, F, Martin, G.S, Kim, S.H. | | Deposit date: | 1997-03-05 | | Release date: | 1998-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of the Ras-interacting domain of RalGDS.
Nat.Struct.Biol., 4, 1997
|
|
6PZI
 
 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP only | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8 | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
6PZB
 
 | |
1FNN
 
 | | CRYSTAL STRUCTURE OF CDC6P FROM PYROBACULUM AEROPHILUM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6, MAGNESIUM ION | | Authors: | Liu, J, Smith, C.L, DeRyckere, D, DeAngelis, K, Martin, G.S, Berger, J.M. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control.
Mol.Cell, 6, 2000
|
|
6PZA
 
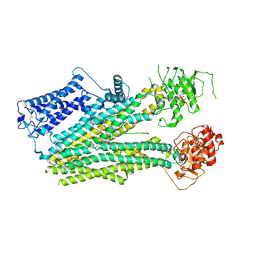 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP and glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
6PZ9
 
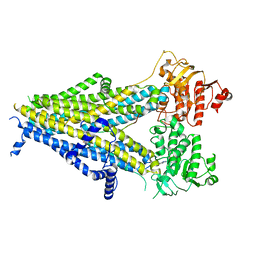 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP and repaglinide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ATP-sensitive inward rectifier potassium channel 11, ... | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
1R5E
 
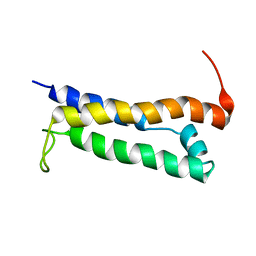 | | Solution structure of the folded core of Pseudomonas syringae effector protein, AvrPto | | Descriptor: | avirulence protein | | Authors: | Wulf, J, Pascuzzi, P.E, Martin, G.B, Nicholson, L.K. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-19 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | The solution structure of type III effector protein AvrPto reveals conformational and dynamic features important for plant pathogenesis.
STRUCTURE, 12, 2004
|
|
