6XK0
 
 | | Albumin-dexamethasone complex | | 分子名称: | Albumin, CITRATE ANION, DEXAMETHASONE, ... | | 著者 | Czub, M.P, Majorek, K.A, Shabalin, I.G, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2020-06-24 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular determinants of vascular transport of dexamethasone in COVID-19 therapy.
Iucrj, 7, 2020
|
|
7CDZ
 
 | |
7CE0
 
 | |
6XHL
 
 | | Covalent complex of SARS-CoV main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 分子名称: | 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | 登録日 | 2020-06-19 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.471 Å) | | 主引用文献 | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHO
 
 | | Covalent complex of SARS-CoV main protease with ethyl (4R)-4-({N-[(4-methoxy-1H-indol-2-yl)carbonyl]-L-leucyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase, ethyl (2E,4S)-4-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | 著者 | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | 登録日 | 2020-06-19 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.446 Å) | | 主引用文献 | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHN
 
 | | Covalent complex of SARS-CoV main protease with 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | 分子名称: | (3S)-3-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-2-oxo-4-[(3S)-2-oxopyrrolidin-3-yl]butyl 2-cyanobenzoate, 1,2-ETHANEDIOL, 3C-like proteinase | | 著者 | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | 登録日 | 2020-06-19 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.377 Å) | | 主引用文献 | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHU
 
 | |
6XHM
 
 | | Covalent complex of SARS-CoV-2 main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | 登録日 | 2020-06-19 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.406 Å) | | 主引用文献 | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6ZFO
 
 | | Association of two complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-17 | | 公開日 | 2020-07-08 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6XGC
 
 | |
6XG3
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, at room temperature | | 分子名称: | CHLORIDE ION, Non-structural protein 3, PHOSPHATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-16 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
6ZER
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with EY6A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | 著者 | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-16 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6XFN
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW243 | | 分子名称: | 3C-like proteinase, GLYCEROL, UAW243 | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-06-15 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-12-02 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-12-02 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6ZDH
 
 | | SARS-CoV-2 Spike glycoprotein in complex with a neutralizing antibody EY6A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-14 | | 公開日 | 2020-07-01 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6XEY
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-4 | | 分子名称: | 2-4 Heavy Chain, 2-4 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Rapp, M, Shapiro, L, Ho, D.D. | | 登録日 | 2020-06-14 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.
Nature, 584, 2020
|
|
6XEZ
 
 | | Structure of SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | 著者 | Chen, J, Malone, B, Llewellyn, E.C, Campbell, E.A, Darst, S.A. | | 登録日 | 2020-06-14 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex.
Cell, 182, 2020
|
|
6ZDG
 
 | | Association of three complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-14 | | 公開日 | 2020-07-29 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7CBT
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Shi, Y, Peng, G. | | 登録日 | 2020-06-13 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.346 Å) | | 主引用文献 | The preclinical inhibitor GS441524 in combination with GC376 efficaciously inhibited the proliferation of SARS-CoV-2 in the mouse respiratory tract.
Emerg Microbes Infect, 10, 2021
|
|
6ZCT
 
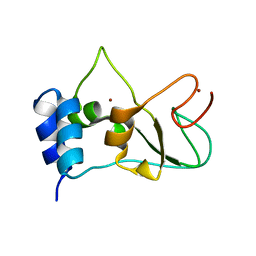 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | 分子名称: | ZINC ION, nsp10 | | 著者 | Rogstam, A, Nyblom, M, Christensen, S, Sele, C, Lindvall, T, Rasmussen, A.A, Andre, I, Fisher, S.Z, Knecht, W, Kozielski, F. | | 登録日 | 2020-06-12 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
6ZCZ
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in ternary complex with EY6A Fab and a nanobody. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EY6A heavy chain, ... | | 著者 | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-12 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZCO
 
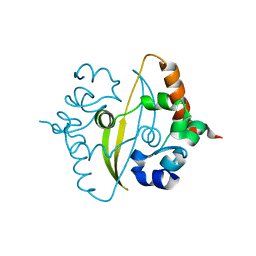 | | Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2, crystal form II | | 分子名称: | Nucleoprotein | | 著者 | Zinzula, L, Basquin, J, Nagy, I, Bracher, A. | | 登録日 | 2020-06-11 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.361 Å) | | 主引用文献 | High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2.
Biochem.Biophys.Res.Commun., 538, 2021
|
|
6XE1
 
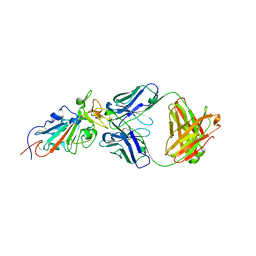 | |
7CB7
 
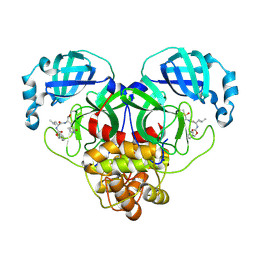 | | 1.7A resolution structure of SARS-CoV-2 main protease (Mpro) in complex with broad-spectrum coronavirus protease inhibitor GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y, Hung, M.C. | | 登録日 | 2020-06-10 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
