5TKP
 
 | |
3UF3
 
 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical isolate PR20 | | 分子名称: | GLYCEROL, HIV-1 protease, YTTRIUM ION | | 著者 | Agniswamy, J, Chen-Hsiang, S, Aniana, A, Sayer, J.M, Louis, J.M, Weber, I.T. | | 登録日 | 2011-10-31 | | 公開日 | 2012-03-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements.
Biochemistry, 51, 2012
|
|
3EL9
 
 | | Crystal structure of atazanavir (ATV) in complex with a multidrug HIV-1 protease (V82T/I84V) | | 分子名称: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | 著者 | Prabu-Jeyabalan, M, King, N, Schiffer, C. | | 登録日 | 2008-09-21 | | 公開日 | 2009-09-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
6DH2
 
 | | Crystal structure of HIV-1 Protease NL4-3 I84V Mutant in complex with UMass6 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | 著者 | Lockbaum, G.J, Schiffer, C.A. | | 登録日 | 2018-05-18 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.978 Å) | | 主引用文献 | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH4
 
 | | Crystal structure of HIV-1 Protease NL4-3 V82I Mutant in complex with UMass1 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | 著者 | Lockbaum, G.J, Schiffer, C.A. | | 登録日 | 2018-05-18 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.943 Å) | | 主引用文献 | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
4ZA9
 
 | | Structure of A. niger fdc1 in complex with a phenylpyruvate derived adduct to the prenylated flavin cofactor | | 分子名称: | 1-deoxy-5-O-phosphono-1-[(1S)-3,3,4,5-tetramethyl-9,11-dioxo-1-(phenylacetyl)-2,3,8,9,10,11-hexahydro-1H,7H-quinolino[1 ,8-fg]pteridin-7-yl]-D-ribitol, MANGANESE (II) ION, POTASSIUM ION, ... | | 著者 | Payne, K.A.P, Leys, D. | | 登録日 | 2015-04-13 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.01 Å) | | 主引用文献 | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
8D0P
 
 | | Human FUT9, unliganded | | 分子名称: | 1,2-ETHANEDIOL, 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9, SULFATE ION, ... | | 著者 | Kadirvelraj, R, Wood, Z.A. | | 登録日 | 2022-05-26 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Structural basis for Lewis antigen synthesis by the alpha 1,3-fucosyltransferase FUT9.
Nat.Chem.Biol., 19, 2023
|
|
8D0U
 
 | | Human FUT9 bound to GDP | | 分子名称: | 1,2-ETHANEDIOL, 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Kadirvelraj, R, Wood, Z.A. | | 登録日 | 2022-05-26 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Structural basis for Lewis antigen synthesis by the alpha 1,3-fucosyltransferase FUT9.
Nat.Chem.Biol., 19, 2023
|
|
8D0R
 
 | | Human FUT9 bound to GDP and H-Type 2 | | 分子名称: | 1,2-ETHANEDIOL, 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Kadirvelraj, R, Wood, Z.A. | | 登録日 | 2022-05-26 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis for Lewis antigen synthesis by the alpha 1,3-fucosyltransferase FUT9.
Nat.Chem.Biol., 19, 2023
|
|
5GTZ
 
 | |
4ZGG
 
 | |
3VC4
 
 | |
4ZSW
 
 | | Pig Brain GABA-AT inactivated by (E)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid | | 分子名称: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | 著者 | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | 登録日 | 2015-05-14 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
2VLA
 
 | | Crystal structure of restriction endonuclease BpuJI recognition domain in complex with cognate DNA | | 分子名称: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 5'-D(*GP*GP*TP*AP*CP*CP*CP*GP*TP*GP *GP*A)-3', 5'-D(*TP*CP*CP*AP*CP*GP*GP*GP*TP*AP *CP*C)-3', ... | | 著者 | Sukackaite, R, Grazulis, S, Bochtler, M, Siksnys, V. | | 登録日 | 2008-01-11 | | 公開日 | 2008-05-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The Recognition Domain of the Bpuji Restriction Endonuclease in Complex with Cognate DNA at 1.3-A Resolution.
J.Mol.Biol., 378, 2008
|
|
4ZSY
 
 | | Pig Brain GABA-AT inactivated by (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid. | | 分子名称: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | 著者 | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | 登録日 | 2015-05-14 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
4ZA4
 
 | | Structure of A. niger Fdc1 with the prenylated-flavin cofactor in the iminium form. | | 分子名称: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Fdc1, MANGANESE (II) ION, ... | | 著者 | Payne, K.A.P, Leys, D. | | 登録日 | 2015-04-13 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
3UCB
 
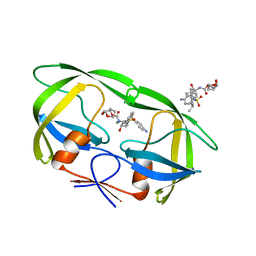 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20 in Complex with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | 著者 | Agniswamy, J, Chen-Hsiang, S, Aniana, A, Sayer, J.M, Louis, J.M, Weber, I.T. | | 登録日 | 2011-10-26 | | 公開日 | 2012-03-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements.
Biochemistry, 51, 2012
|
|
3F0T
 
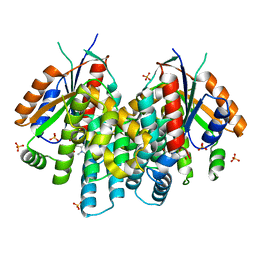 | | Crystal structure of thymidine kinase from Herpes simplex virus type 1 in complex with N-methyl-DHBT | | 分子名称: | N-Methyl-6-(1,3-dihydroxy-isobutyl)thymine, SULFATE ION, Thymidine kinase | | 著者 | Pernot, L, Perozzo, R, Westermaier, Y, Martic, M, Ametamey, S, Scapozza, L. | | 登録日 | 2008-10-26 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Synthesis, crystal structure, and in vitro biological evaluation of C-6 pyrimidine derivatives: new lead structures for monitoring gene expression in vivo.
Nucleosides Nucleotides Nucleic Acids, 30, 2011
|
|
5ES2
 
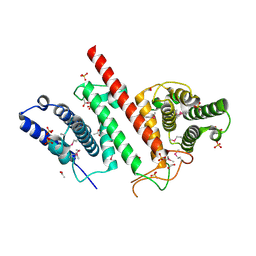 | | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | 著者 | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-11-16 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To Be Published
|
|
4YOA
 
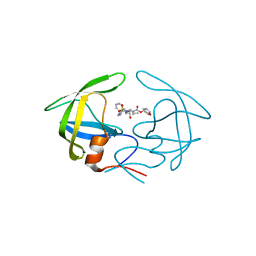 | | Crsystal structure HIV-1 Protease MDR769 L33F Complexed with darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 Protease | | 著者 | Kuiper, B.D, Keusch, B, Dewdney, T.G, Chordia, P, Brunzelle, J.S, Ross, K, Kovari, I.A, MacArthur, R, Salimnia, H, Kovari, L.C. | | 登録日 | 2015-03-11 | | 公開日 | 2015-07-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.697 Å) | | 主引用文献 | The L33F darunavir resistance mutation acts as a molecular anchor reducing the flexibility of the HIV-1 protease 30s and 80s loops.
Biochem Biophys Rep, 2, 2015
|
|
3VBQ
 
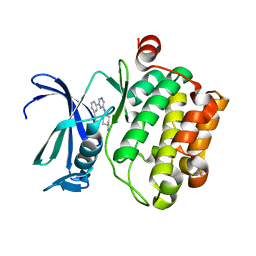 | |
3F6T
 
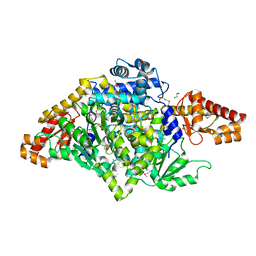 | |
7V6G
 
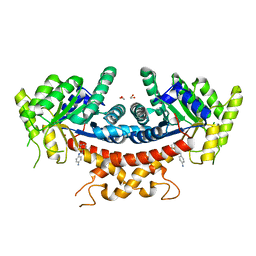 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase mutation C157S with CN39 | | 分子名称: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION, ... | | 著者 | Cao, H, Huang, Y, Chen, H, Wan, C, Ren, Y, Wan, J. | | 登録日 | 2021-08-20 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.343 Å) | | 主引用文献 | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
3CYX
 
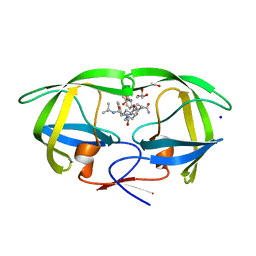 | | Crystal structure of HIV-1 mutant I50V and inhibitor saquinavira | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, GLYCEROL, ... | | 著者 | Liu, F, Weber, I.T. | | 登録日 | 2008-04-27 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
5YDB
 
 | | Crystal structure of the complex of type II dehydroquinate dehydratase from Acinetobacter baumannii with dehydroquinic acid at 1.76 Angstrom resolution | | 分子名称: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase, SODIUM ION | | 著者 | Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2017-09-12 | | 公開日 | 2017-09-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of the complex of type II dehydroquinate dehydratase from Acinetobacter baumannii with dehydroquinic acid at 1.76 Angstrom resolution
To Be Published
|
|
