7KYT
 
 | | The aminoacrylate form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, cesium ion at the metal coordination site and benzimidazole (BZI) at the enzyme beta-site at 1.35 Angstrom resolution | | 分子名称: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | 著者 | Hilario, E, Dunn, M.F, Mueller, L.J. | | 登録日 | 2020-12-08 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | The aminoacrylate form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, cesium ion at the metal coordination site and benzimidazole (BZI) at the enzyme beta-site at 1.35 Angstrom resolution.
To be Published
|
|
7L03
 
 | |
7L1H
 
 | |
7LEV
 
 | |
7LGX
 
 | | The aminoacrylate form of mutant beta-K167T Salmonella typhimurium Tryptophan Synthase in complex with with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, benzimidazole (BZI) at the enzyme beta site and cesium ion at the metal coordination site at 1.80 Angstrom resolution | | 分子名称: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | 著者 | Hilario, E, Dunn, M.F, Mueller, L.J. | | 登録日 | 2021-01-21 | | 公開日 | 2022-01-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The aminoacrylate form of mutant beta-K167T Salmonella typhimurium Tryptophan Synthase in complex with with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, benzimidazole (BZI) at the enzyme beta site and cesium ion at the metal coordination site at 1.80 Angstrom resolution.
To be Published
|
|
1ESL
 
 | |
7MBV
 
 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 5 mM calcium and 0.5 mM NDNA | | 分子名称: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ruan, Z, Lu, W, Du, J, Haley, E. | | 登録日 | 2021-04-01 | | 公開日 | 2021-07-07 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5VWR
 
 | | E.coli Aspartate aminotransferase-(1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid (FCP)-alpha-ketoglutarate | | 分子名称: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, Aspartate aminotransferase, GLYCEROL | | 著者 | Mascarenhas, R, Liu, D, Le, H, Silverman, R. | | 登録日 | 2017-05-22 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Selective Targeting by a Mechanism-Based Inactivator against Pyridoxal 5'-Phosphate-Dependent Enzymes: Mechanisms of Inactivation and Alternative Turnover.
Biochemistry, 56, 2017
|
|
7LXJ
 
 | |
7LT4
 
 | |
3GHQ
 
 | | Crystal Structure of E. coli W35F BFR mutant | | 分子名称: | Bacterioferritin, FE (III) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Crow, A, Lawson, T.L, Lewin, A, Moore, G.R, Le Brun, N.E. | | 登録日 | 2009-03-04 | | 公開日 | 2009-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Monitoring the iron status of the ferroxidase center of Escherichia coli bacterioferritin using fluorescence spectroscopy.
Biochemistry, 48, 2009
|
|
7L4L
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C8-crypto Acyl Carrier Protein, AcpP | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, SODIUM ION, ... | | 著者 | Mindrebo, J.T, Chen, A, Kim, W.E, Burkart, M.D, Noel, J.P. | | 登録日 | 2020-12-19 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure and Mechanistic Analyses of the Gating Mechanism of Elongating Ketosynthases
Acs Catalysis, 11, 2021
|
|
7B9H
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-42a | | 分子名称: | 1,2-ETHANEDIOL, 3-[(~{E})-1-(3-cyclopentyloxy-4-methoxy-phenyl)ethylideneamino]oxy-1-morpholin-4-yl-propan-1-one, MAGNESIUM ION, ... | | 著者 | Torretta, A, Abbate, S, Parisini, E. | | 登録日 | 2020-12-14 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Design, synthesis, biological evaluation and structural characterization of novel GEBR library PDE4D inhibitors.
Eur.J.Med.Chem., 223, 2021
|
|
1ZL6
 
 | | Crystal structure of Tyr350Ala mutant of Clostridium botulinum neurotoxin E catalytic domain | | 分子名称: | SULFATE ION, ZINC ION, botulinum neurotoxin type E | | 著者 | Agarwal, R, Binz, T, Swaminathan, S. | | 登録日 | 2005-05-05 | | 公開日 | 2005-06-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
4Z3N
 
 | |
1EZF
 
 | | CRYSTAL STRUCTURE OF HUMAN SQUALENE SYNTHASE | | 分子名称: | FARNESYL-DIPHOSPHATE FARNESYLTRANSFERASE, N-{2-[TRANS-7-CHLORO-1-(2,2-DIMETHYL-PROPYL) -5-NAPHTHALEN-1-YL-2-OXO-1,2,3,5-TETRAHYDRO-BENZO[E] [1,4]OXAZEPIN-3-YL]-ACETYL}-ASPARTIC ACID | | 著者 | Pandit, J, Danley, D.E, Schulte, G.K, Mazzalupo, S.M, Pauly, T.A, Hayward, C.M, Hamanaka, E.S, Thompson, J.F, Harwood, H.J. | | 登録日 | 2000-05-10 | | 公開日 | 2000-10-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis.
J.Biol.Chem., 275, 2000
|
|
1F00
 
 | | CRYSTAL STRUCTURE OF C-TERMINAL 282-RESIDUE FRAGMENT OF ENTEROPATHOGENIC E. COLI INTIMIN | | 分子名称: | INTIMIN | | 著者 | Luo, Y, Frey, E.A, Pfuetzner, R.A, Creagh, A.L, Knoechel, D.G, Haynes, C.A, Finlay, B.B, Strynadka, N.C.J. | | 登録日 | 2000-05-12 | | 公開日 | 2000-07-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex.
Nature, 405, 2000
|
|
4O4R
 
 | | Murine Norovirus RdRp in complex with PPNDS | | 分子名称: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, RNA-dependent-RNA-polymerase, SULFATE ION | | 著者 | Croci, R, Tarantino, D, Milani, M, Pezzullo, M, Bolognesi, M, Mastrangelo, E. | | 登録日 | 2013-12-19 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | PPNDS inhibits murine Norovirus RNA-dependent RNA-polymerase mimicking two RNA stacking bases.
Febs Lett., 588, 2014
|
|
1ZL5
 
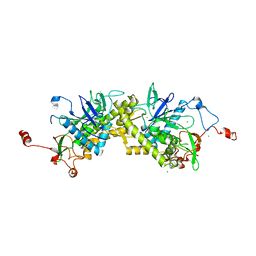 | | Crystal structure of Glu335Gln mutant of Clostridium botulinum neurotoxin E catalytic domain | | 分子名称: | CHLORIDE ION, botulinum neurotoxin type E | | 著者 | Agarwal, R, Binz, T, Swaminathan, S. | | 登録日 | 2005-05-05 | | 公開日 | 2005-07-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
7BH2
 
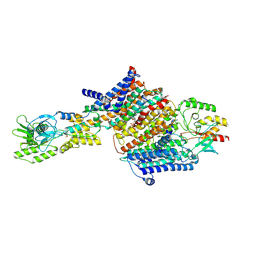 | | Cryo-EM Structure of KdpFABC in E2Pi state with BeF3 and K+ | | 分子名称: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | 著者 | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | 登録日 | 2021-01-09 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1GOS
 
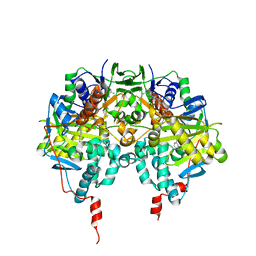 | | Human Monoamine Oxidase B | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE, N-[(E)-METHYL](PHENYL)-N-[(E)-2-PROPENYLIDENE]METHANAMINIUM | | 著者 | Binda, C, Newton-Vinson, P, Hubalek, F, Edmondson, D.E, Mattevi, A. | | 登録日 | 2001-10-26 | | 公開日 | 2001-11-29 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of Human Monoamine Oxidase B, a Drug Target for the Treatment of Neurological Disorders
Nat.Struct.Biol., 9, 2001
|
|
1ZKW
 
 | | Crystal structure of Arg347Ala mutant of botulinum neurotoxin E catalytic domain | | 分子名称: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | 著者 | Agarwal, R, Binz, T, Swaminathan, S. | | 登録日 | 2005-05-04 | | 公開日 | 2005-06-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1Z9H
 
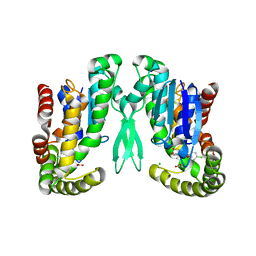 | | Microsomal prostaglandin E synthase type-2 | | 分子名称: | ACETATE ION, CHLORIDE ION, INDOMETHACIN, ... | | 著者 | Yamada, T, Komoto, J, Watanabe, K, Ohmiya, Y, Takusagawa, F. | | 登録日 | 2005-04-02 | | 公開日 | 2005-05-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure and Possible Catalytic Mechanism of Microsomal Prostaglandin E Synthase Type 2 (mPGES-2).
J.Mol.Biol., 348, 2005
|
|
5AQ6
 
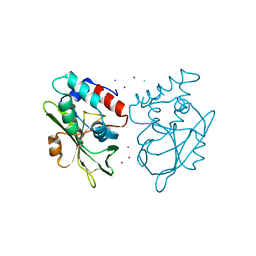 | | Structure of E. coli ZinT at 1.79 Angstrom | | 分子名称: | ACETIC ACID, METAL-BINDING PROTEIN ZINT, ZINC ION | | 著者 | Santo, P.E, Colaco, H.G, Matias, P, Vicente, J.B, Bandeiras, T.M. | | 登録日 | 2015-09-21 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Roles of Escherichia Coli Zint in Cobalt, Mercury and Cadmium Resistance and Structural Insights Into the Metal Binding Mechanism
Metallomics, 8, 2016
|
|
1ZKX
 
 | | Crystal structure of Glu158Ala/Thr159Ala/Asn160Ala- a triple mutant of Clostridium botulinum neurotoxin E catalytic domain | | 分子名称: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | 著者 | Agarwal, R, Binz, T, Swaminathan, S. | | 登録日 | 2005-05-04 | | 公開日 | 2005-07-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
