3DPL
 
 | |
3DQV
 
 | | Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation | | 分子名称: | Cullin-5, NEDD8, Rbx1, ... | | 著者 | Duda, D.M, Borg, L.A, Scott, D.C, Hunt, H.W, Hammel, M, Schulman, B.A. | | 登録日 | 2008-07-09 | | 公開日 | 2008-09-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation.
Cell(Cambridge,Mass.), 134, 2008
|
|
3E0C
 
 | | Crystal Structure of DNA Damage-Binding protein 1(DDB1) | | 分子名称: | DNA damage-binding protein 1 | | 著者 | Amaya, M.F, Xu, L, Hao, H, Bountra, C, Wickstroem, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2008-07-31 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
3EI2
 
 | |
3EI1
 
 | |
3EI4
 
 | |
3EI3
 
 | | Structure of the hsDDB1-drDDB2 complex | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, TETRAETHYLENE GLYCOL | | 著者 | Scrima, A, Thoma, N.H. | | 登録日 | 2008-09-15 | | 公開日 | 2009-01-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
8Q7R
 
 | | Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide | | 分子名称: | 5-azanyl-1-oxidanyl-pentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | 登録日 | 2023-08-16 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
8T9A
 
 | | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3 | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, Melanoma-associated antigen 3 | | 著者 | Duda, D, Righetto, G, Li, Y, Loppnau, P, Seitova, A, Santhakumar, V, Halabelian, L, Yin, Y. | | 登録日 | 2023-06-23 | | 公開日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3
To Be Published
|
|
8TNR
 
 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 2 | | 分子名称: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | 著者 | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | 登録日 | 2023-08-02 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8TNQ
 
 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 1 | | 分子名称: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | 著者 | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | 登録日 | 2023-08-02 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (2.41 Å) | | 主引用文献 | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | 分子名称: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | 登録日 | 2023-07-11 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TNP
 
 | | Cryo-EM structure of DDB1dB:CRBN:Pomalidomide:SD40 | | 分子名称: | DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, Protein cereblon, ... | | 著者 | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | 登録日 | 2023-08-02 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8Q7E
 
 | |
8D7U
 
 | |
8D81
 
 | | Cereblon~DDB1 bound to Pomalidomide | | 分子名称: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | 著者 | Watson, E.R, Lander, G.C. | | 登録日 | 2022-06-07 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8D7Y
 
 | |
8D7W
 
 | |
8D7Z
 
 | |
8CVP
 
 | | Cereblon-DDB1 in the Apo form | | 分子名称: | DNA damage-binding protein 1, Protein cereblon, ZINC ION | | 著者 | Watson, E.R, Lander, G.C. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8D7X
 
 | |
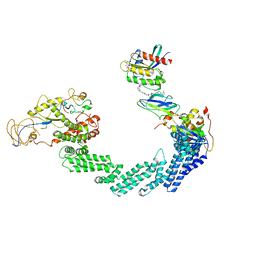
8D7V
 
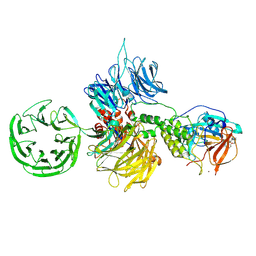 | |
8D80
 
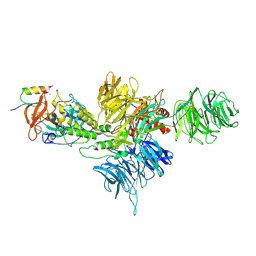 | | Cereblon~DDB1 bound to Iberdomide and Ikaros ZF1-2-3 | | 分子名称: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, DNA-binding protein Ikaros, ... | | 著者 | Watson, E.R, Lander, G.C. | | 登録日 | 2022-06-07 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8QU8
 
 | | PROTAC-mediated complex of KRAS with VHL/Elongin-B/Elongin-C/Cullin-2/Rbx1 | | 分子名称: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Fischer, G, Peter, D, Arce-Solano, S. | | 登録日 | 2023-10-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Targeting cancer with small molecule pan-KRAS degraders
Biorxiv, 2023
|
|
8RHZ
 
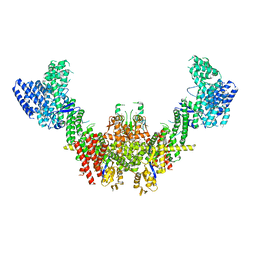 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer | | 分子名称: | Cullin-9, E3 ubiquitin-protein ligase RBX1, ZINC ION | | 著者 | Hopf, L.V.M, Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2023-12-17 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
