8AJN
 
 | | Structure of the human DDB1-DCAF12 complex | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1 | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJM
 
 | | Structure of human DDB1-DCAF12 in complex with the C-terminus of CCT5 | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJO
 
 | | Negative-stain electron microscopy structure of DDB1-DCAF12-CCT5 | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (30.6 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8OR2
 
 | | CAND1-CUL1-RBX1-DCNL1 | | 分子名称: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, DCN1-like protein 1, ... | | 著者 | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | 登録日 | 2023-04-12 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
4XR8
 
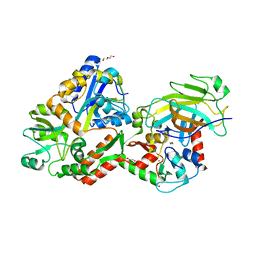 | | Crystal structure of the HPV16 E6/E6AP/p53 ternary complex at 2.25 A resolution | | 分子名称: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Martinez-Zapien, D, Ruiz, F.X, Mitschler, A, Podjarny, A, Trave, G, Zanier, K. | | 登録日 | 2015-01-20 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53.
Nature, 529, 2016
|
|
1HXD
 
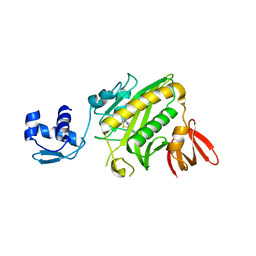 | | CRYSTAL STRUCTURE OF E. COLI BIOTIN REPRESSOR WITH BOUND BIOTIN | | 分子名称: | BIOTIN, BIRA BIFUNCTIONAL PROTEIN | | 著者 | Kwon, K, Streaker, E.D, Ruparelia, S, Beckett, D. | | 登録日 | 2001-01-12 | | 公開日 | 2001-05-30 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Corepressor-induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6WCQ
 
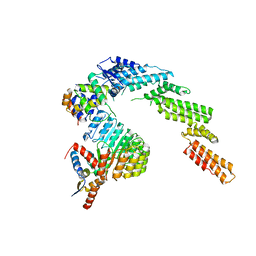 | | Structure of a substrate-bound DQC ubiquitin ligase | | 分子名称: | Cullin-1, F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, ... | | 著者 | Mena, E.L, Jevtic, P, Greber, B.J, Gee, C.L, Lew, B.G, Akopian, D, Nogales, E, Kuriyan, J, Rape, M. | | 登録日 | 2020-03-31 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (8.5 Å) | | 主引用文献 | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
6GYS
 
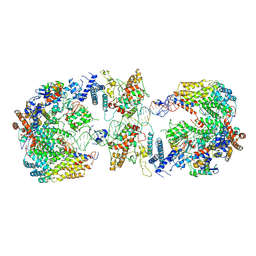 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYU
 
 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-02 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYP
 
 | | Cryo-EM structure of the CBF3-core-Ndc10-DBD complex of the budding yeast kinetochore | | 分子名称: | ARGININE, ASPARAGINE, Centromere DNA-binding protein complex CBF3 subunit A, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6BN8
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET55 PROTAC. | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.990035 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BN9
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET70 PROTAC | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (4.382 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BN7
 
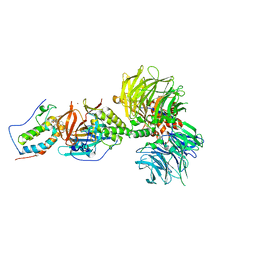 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET23 PROTAC. | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.501 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BNB
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET57 PROTAC | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Ishoey, M, He, Z, Zhang, T, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (6.343 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BOY
 
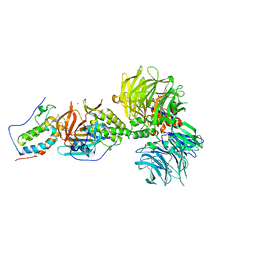 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET6 PROTAC. | | 分子名称: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(8-{[({2-[(3S)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}oxy)acetyl]amino}octyl)acetamide, Bromodomain-containing protein 4, DNA damage-binding protein 1, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-21 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.33 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
9FGQ
 
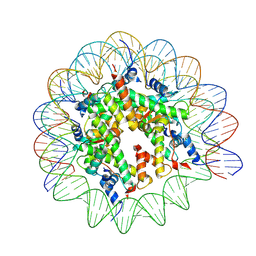 | | Structure of human APC3loop 375-381 bound to the NCP | | 分子名称: | Cell division cycle protein 27 homolog, DNA (131-MER), DNA (132-MER), ... | | 著者 | Young, R.V.C, Muhammad, R, Alfieri, C. | | 登録日 | 2024-05-24 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Spatial control of the APC/C ensures the rapid degradation of cyclin B1.
Embo J., 43, 2024
|
|
6F07
 
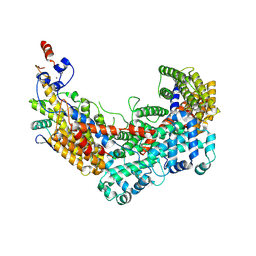 | | CBF3 Core Complex | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit B, Suppressor of kinetochore protein 1, ZINC ION | | 著者 | Leber, V, Singleton, M.R. | | 登録日 | 2017-11-17 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for assembly of the CBF3 kinetochore complex.
EMBO J., 37, 2018
|
|
7ZN7
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and rat STAT2 CCD | | 分子名称: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | 著者 | Lauer, S, Spahn, C.M.T, Schwefel, D. | | 登録日 | 2022-04-20 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
7ZNN
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and full-length rat STAT2 | | 分子名称: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | 著者 | Lauer, S, Spahn, C.M.T, Schwefel, D. | | 登録日 | 2022-04-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
5XIS
 
 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form I | | 分子名称: | E3 ubiquitin-protein ligase RNF168, MAGNESIUM ION, Ubiquitin-40S ribosomal protein S27a, ... | | 著者 | Takahashi, T.S, Sato, Y, Fukai, S. | | 登録日 | 2017-04-27 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
7MEY
 
 | | Structure of yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron | | 分子名称: | 2-(ethylamino)ethane-1-thiol, E3 ubiquitin-protein ligase UBR1, Monoubiquitinated N-degron, ... | | 著者 | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-04-08 | | 公開日 | 2021-11-24 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
8C07
 
 | |
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | 著者 | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | 登録日 | 2023-12-15 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | 分子名称: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | 登録日 | 2023-04-26 | | 公開日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
1FLO
 
 | | FLP Recombinase-Holliday Junction Complex I | | 分子名称: | FLP RECOMBINASE, PHOSPHONIC ACID, SYMMETRIZED FRT DNA SITES | | 著者 | Chen, Y, Narendra, U, Iype, L.E, Cox, M.M, Rice, P.A. | | 登録日 | 2000-08-14 | | 公開日 | 2000-09-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping.
Mol.Cell, 6, 2000
|
|
