9BRD
 
 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3 | | 分子名称: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Coupland, C.E, Rubinstein, J.L. | | 登録日 | 2024-05-11 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9CMS
 
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease drug resistant mutant (E166V) in complex with ensitrelvir (ESV) | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Kovalevsky, A, Coates, L, Gerlits, O. | | 登録日 | 2024-07-15 | | 公開日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Effects of SARS-CoV-2 Main Protease Mutations at Positions L50, E166, and L167 Rendering Resistance to Covalent and Noncovalent Inhibitors.
J.Med.Chem., 2024
|
|
8YG6
 
 | | The pre-fusion structure of baculovirus fusion protein GP64 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Major envelope glycoprotein | | 著者 | Du, D, Guo, J, Li, S. | | 登録日 | 2024-02-26 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structural transition of GP64 triggered by a pH-sensitive multi-histidine switch.
Nat Commun, 15, 2024
|
|
9CMN
 
 | |
9AU1
 
 | | SARS-CoV-2 XBB.1.5 RBD bound to the VIR-7229 and the S309 Fab fragments | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Rietz, T, Park, Y.J, Errico, J, Czudnochowski, N, Nix, J.C, Corti, D, Snell, G, Marco, A.D, Pinto, D, Cameroni, E, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D, Structural Genomics Consortium (SGC) | | 登録日 | 2024-02-27 | | 公開日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 2024
|
|
9C82
 
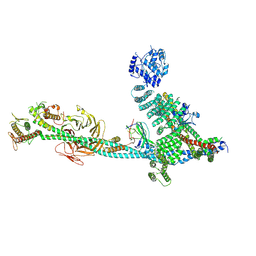 | | Structure of human ULK1C:PI3KC3-C1 supercomplex | | 分子名称: | Beclin 1-associated autophagy-related key regulator, Beclin-1, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | 著者 | Chen, M, Hurley, J.H. | | 登録日 | 2024-06-11 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (6.84 Å) | | 主引用文献 | Structure and activation of the human autophagy-initiating ULK1C:PI3KC3-C1 supercomplex
bioRxiv, 2023
|
|
9BRQ
 
 | | Intact V-ATPase State 3 and synaptophysin complex in mouse brain isolated synaptic vesicles | | 分子名称: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | 著者 | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | 登録日 | 2024-05-11 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
6WVG
 
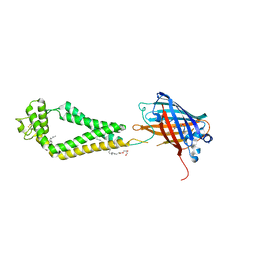 | | human CD53 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Green fluorescent protein, ... | | 著者 | Yang, Y, Liu, S, Li, W. | | 登録日 | 2020-05-06 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Open conformation of tetraspanins shapes interaction partner networks on cell membranes.
Embo J., 39, 2020
|
|
9BRC
 
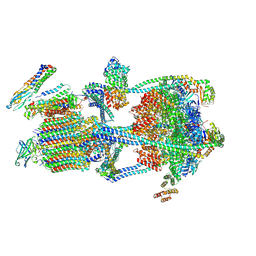 | |
9BEI
 
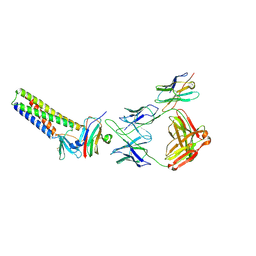 | |
9BQ0
 
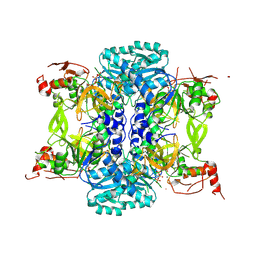 | |
9CMJ
 
 | |
9CMU
 
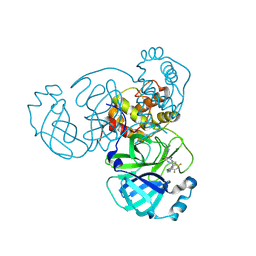 | | Room-temperature X-ray structure of SARS-CoV-2 main protease drug resistant mutant (L50F, E166V) in complex with ensitrelvir (ESV) | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Kovalevsky, A, Coates, L, Gerlits, O. | | 登録日 | 2024-07-15 | | 公開日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Effects of SARS-CoV-2 Main Protease Mutations at Positions L50, E166, and L167 Rendering Resistance to Covalent and Noncovalent Inhibitors.
J.Med.Chem., 2024
|
|
9IZN
 
 | | Crystal structure of HKU1A RBD bound to TMPRSS2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, Transmembrane protease serine 2 | | 著者 | Wang, W, Xu, Y, Zhang, S. | | 登録日 | 2024-08-01 | | 公開日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | High-resolution crystal structure of human coronavirus HKU1 receptor binding domain bound to TMPRSS2 receptor
Hlife, 2024
|
|
9B8O
 
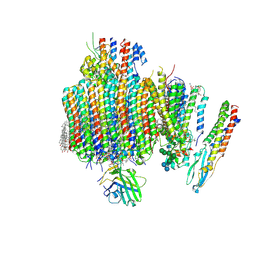 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, Vo | | 分子名称: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Coupland, C.E, Rubinstein, J.L. | | 登録日 | 2024-03-31 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9ATM
 
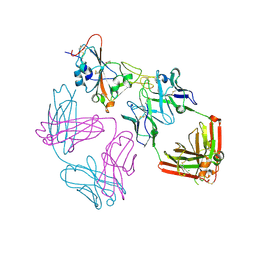 | | SARS-CoV-2 EG.5 RBD bound to the VIR-7229 and the S2H97 Fab fragments | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Rietz, T, Park, Y.J, Errico, J, Czudnochowski, N, Nix, J.C, Corti, D, Snell, G, Marco, A.D, Pinto, D, Cameroni, E, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | 登録日 | 2024-02-27 | | 公開日 | 2024-10-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 2024
|
|
9BRZ
 
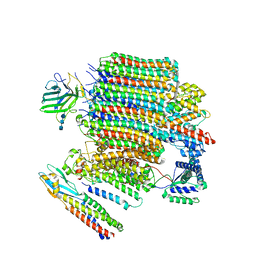 | | V0-only V-ATPase and synaptophysin complex in mouse brain isolated synaptic vesicles | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | 登録日 | 2024-05-12 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
6YQJ
 
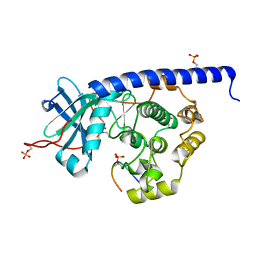 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with open-chain Fasudil-derivative 2-[isoquinolin-5-ylsulfonyl(propyl)amino]ethylazanium (soaked) | | 分子名称: | 2-[isoquinolin-5-ylsulfonyl(propyl)amino]ethylazanium, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | 著者 | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | 登録日 | 2020-04-17 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6Y8Y
 
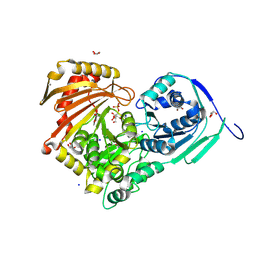 | | Structure of Baltic Herring (Clupea Harengus) Phosphoglucomutase 5 (PGM5) with bound Glucose-1-Phosphate | | 分子名称: | 1-O-phosphono-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | 著者 | Gustafsson, R, Eckhard, U, Selmer, M. | | 登録日 | 2020-03-06 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure and Characterization of Phosphoglucomutase 5 from Atlantic and Baltic Herring-An Inactive Enzyme with Intact Substrate Binding.
Biomolecules, 10, 2020
|
|
6YNT
 
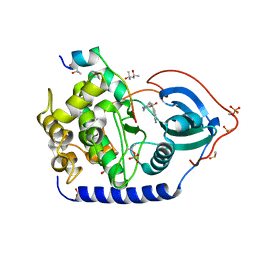 | | Crystal structure of the cAMP-dependent protein kinase A in complex with aminofasudil and PKI (5-24) | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1,4-diazepan-1-ylsulfonyl)isoquinolin-1-amine, ... | | 著者 | Oebbeke, M, Gerber, H.-D, Heine, A, Klebe, G. | | 登録日 | 2020-04-14 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6YNA
 
 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with Fasudil (M77, soaked) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | 著者 | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | 登録日 | 2020-04-13 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6YQK
 
 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with a methylisoquinoline Fasudil-derivative (soaked) | | 分子名称: | 5-(1,4-diazepan-1-ylsulfonyl)-4-methyl-isoquinoline, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | 著者 | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | 登録日 | 2020-04-17 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6YPS
 
 | | Crystal structure of the cAMP-dependent protein kinase A in complex with 4-hydroxybenzamidine | | 分子名称: | 4-oxidanylbenzenecarboximidamide, DIMETHYL SULFOXIDE, cAMP-dependent protein kinase catalytic subunit alpha | | 著者 | Oebbeke, M, Siefker, C, Heine, A, Klebe, G. | | 登録日 | 2020-04-16 | | 公開日 | 2020-12-09 | | 最終更新日 | 2020-12-30 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6YNB
 
 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with short-chain Fasudil-derivative N-(2-aminoethyl)isoquinoline-5-sulfonamide (soaked) | | 分子名称: | DIMETHYL SULFOXIDE, N-(2-AMINOETHYL)ISOQUINOLINE-5-SULFONAMIDE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | 著者 | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | 登録日 | 2020-04-13 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6YNR
 
 | | Crystal structure of the cAMP-dependent protein kinase A in complex with 1,7-Naphthyridin-8-amine (soaked) and PKI (5-24) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,7-naphthyridin-8-amine, DIMETHYL SULFOXIDE, ... | | 著者 | Oebbeke, M, Heine, A, Klebe, G. | | 登録日 | 2020-04-14 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
