3IT7
 
 | | Crystal Structure of the LasA virulence factor from Pseudomonas aeruginosa | | 分子名称: | GLYCEROL, L(+)-TARTARIC ACID, Protease lasA, ... | | 著者 | Spencer, J, Murphy, L.M, Conners, R, Sessions, R.B, Gamblin, S.J. | | 登録日 | 2009-08-27 | | 公開日 | 2009-11-17 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Crystal structure of the LasA virulence factor from Pseudomonas aeruginosa: substrate specificity and mechanism of M23 metallopeptidases.
J.Mol.Biol., 396, 2010
|
|
3IT8
 
 | |
3IT9
 
 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in apo state | | 分子名称: | D-alanyl-D-alanine carboxypeptidase dacC, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | 登録日 | 2009-08-27 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
3ITA
 
 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in acyl-enzyme complex with ampicillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, D-alanyl-D-alanine carboxypeptidase dacC, ... | | 著者 | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | 登録日 | 2009-08-27 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
3ITB
 
 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in complex with a substrate fragment | | 分子名称: | D-alanyl-D-alanine carboxypeptidase DacC, Peptidoglycan substrate (AMV)A(FGA)K(DAL)(DAL), SULFATE ION, ... | | 著者 | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | 登録日 | 2009-08-27 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
3ITC
 
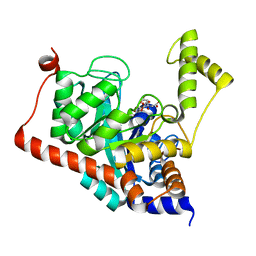 | | Crystal structure of Sco3058 with bound citrate and glycerol | | 分子名称: | CITRIC ACID, GLYCEROL, ZINC ION, ... | | 著者 | Nguyen, T.T, Cummings, J.A, Tsai, C.-L, Barondeau, D.P, Raushel, F.M. | | 登録日 | 2009-08-28 | | 公開日 | 2010-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase
Biochemistry, 49, 2010
|
|
3ITD
 
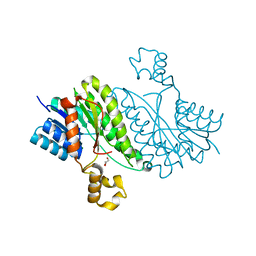 | |
3ITE
 
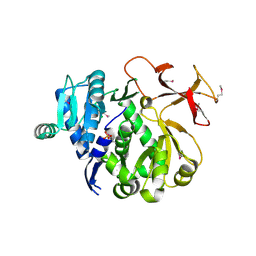 | | The third adenylation domain of the fungal SidN non-ribosomal peptide synthetase | | 分子名称: | CHLORIDE ION, SULFATE ION, SidN siderophore synthetase | | 著者 | Lee, T.V, Lott, J.S, Johnson, R.D, Johnson, L.J, Arcus, V.L. | | 登録日 | 2009-08-28 | | 公開日 | 2009-11-17 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a eukaryotic nonribosomal peptide synthetase adenylation domain that activates a large hydroxamate amino acid in siderophore biosynthesis
J.Biol.Chem., 285, 2010
|
|
3ITF
 
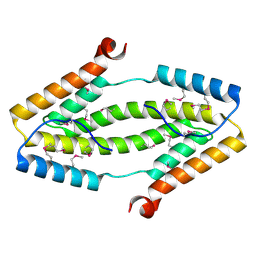 | |
3ITG
 
 | |
3ITH
 
 | | Crystal structure of the HIV-1 reverse transcriptase bound to a 6-vinylpyrimidine inhibitor | | 分子名称: | 6-ethenyl-N,N-dimethyl-2-(methylsulfonyl)pyrimidin-4-amine, Reverse transcriptase/ribonuclease H, p51 RT | | 著者 | Freisz, S, Bec, G, Wolff, P, Dumas, P, Radi, M, Botta, M. | | 登録日 | 2009-08-28 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of HIV-1 Reverse Transcriptase Bound to a Non-Nucleoside Inhibitor with a Novel Mechanism of Action
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3ITI
 
 | | Structure of bovine trypsin with the MAD triangle B3C | | 分子名称: | 5-amino-2,4,6-tribromobenzene-1,3-dicarboxylic acid, BENZAMIDINE, CALCIUM ION, ... | | 著者 | Beck, T, da Cunha, C.E, Sheldrick, G.M. | | 登録日 | 2009-08-28 | | 公開日 | 2009-10-27 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | How to get the magic triangle and the MAD triangle into your protein crystal.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3ITJ
 
 | | Crystal structure of Saccharomyces cerevisiae thioredoxin reductase 1 (Trr1) | | 分子名称: | CITRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase 1 | | 著者 | Oliveira, M.A, Discola, K.F, Alves, S.V, Medrano, F.J, Guimaraes, B.G, Netto, L.E.S. | | 登録日 | 2009-08-28 | | 公開日 | 2010-03-31 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Insights into the specificity of thioredoxin reductase-thioredoxin interactions. A structural and functional investigation of the yeast thioredoxin system.
Biochemistry, 49, 2010
|
|
3ITK
 
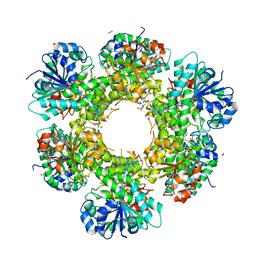 | | Crystal structure of human UDP-glucose dehydrogenase Thr131Ala, apo form. | | 分子名称: | 1,2-ETHANEDIOL, TETRAETHYLENE GLYCOL, UDP-glucose 6-dehydrogenase | | 著者 | Chaikuad, A, Egger, S, Yue, W.W, Sethi, R, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Kavanagh, K.L, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | 登録日 | 2009-08-28 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
3ITL
 
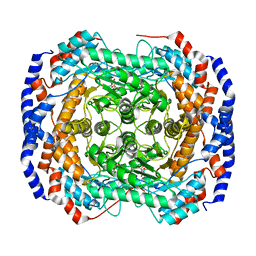 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant D327N in complex with L-rhamnulose | | 分子名称: | 6-deoxy-beta-L-fructofuranose, L-rhamnose isomerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2009-08-28 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITM
 
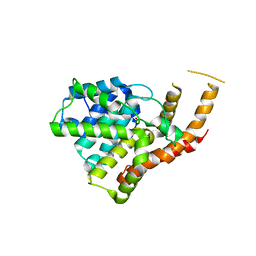 | | Catalytic domain of hPDE2A | | 分子名称: | ZINC ION, cGMP-dependent 3',5'-cyclic phosphodiesterase | | 著者 | Pandit, J. | | 登録日 | 2009-08-28 | | 公開日 | 2009-10-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ITN
 
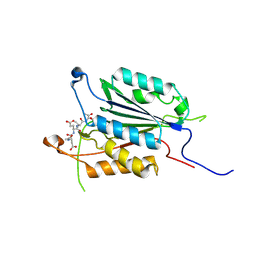 | | Crystal structure of Pseudo-activated Procaspase-3 | | 分子名称: | ACETYL-ASP-GLU-VAL-ASP-CHLOROMETHYL KETONE inhibitor, Caspase-3 | | 著者 | Walters, J, Pop, C, Scott, F.L, Drag, M, Swartz, P.D, Mattos, C, Salvesen, G.S, Clark, A.C. | | 登録日 | 2009-08-28 | | 公開日 | 2010-03-02 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | A constitutively active and uninhibitable caspase-3 zymogen efficiently induces apoptosis.
Biochem.J., 424, 2009
|
|
3ITO
 
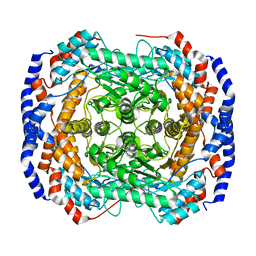 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant D327N in complex with D-psicose | | 分子名称: | L-rhamnose isomerase, MANGANESE (II) ION, alpha-D-psicofuranose | | 著者 | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2009-08-28 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITP
 
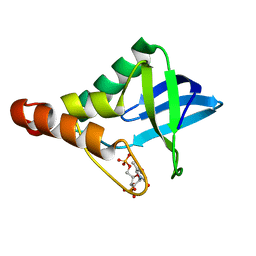 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS F34K at cryogenic temperature | | 分子名称: | CALCIUM ION, Nuclease A, THYMIDINE-3',5'-DIPHOSPHATE | | 著者 | Khangulov, V.S, Schlessman, J.L, Heroux, A, Garcia-Moreno, E.B. | | 登録日 | 2009-08-28 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of Staphylococcal nuclease variant Delta+PHS F34K at cryogenic temperature
To be Published
|
|
3ITQ
 
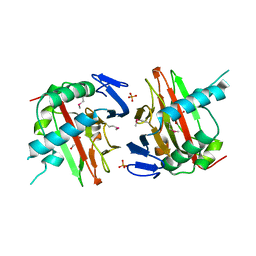 | | Crystal Structure of a Prolyl 4-Hydroxylase from Bacillus anthracis | | 分子名称: | GLYCEROL, PHOSPHATE ION, Prolyl 4-hydroxylase, ... | | 著者 | Culpepper, M.A, Scott, E.E, Limburg, J. | | 登録日 | 2009-08-28 | | 公開日 | 2009-12-15 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of prolyl 4-hydroxylase from Bacillus anthracis.
Biochemistry, 49, 2010
|
|
3ITT
 
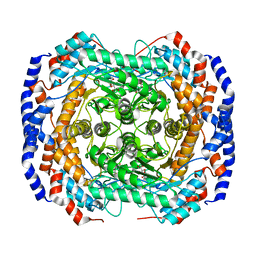 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329K in complex with L-rhamnose | | 分子名称: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2009-08-28 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITU
 
 | | hPDE2A catalytic domain complexed with IBMX | | 分子名称: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, ZINC ION, ... | | 著者 | Pandit, J. | | 登録日 | 2009-08-28 | | 公開日 | 2009-10-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ITV
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329K in complex with D-psicose | | 分子名称: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2009-08-28 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITW
 
 | |
3ITX
 
 | | Mn2+ bound form of Pseudomonas stutzeri L-rhamnose isomerase | | 分子名称: | L-rhamnose isomerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2009-08-28 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
