8KHP
 
 | | CULLIN3-KLHL22-RBX1 E3 ligase | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch-like protein 22 | | 著者 | Su, M.-Y, Su, M.-Y. | | 登録日 | 2023-08-22 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
5FQD
 
 | | Structural basis of Lenalidomide induced CK1a degradation by the crl4crbn ubiquitin ligase | | 分子名称: | CASEIN KINASE I ISOFORM ALPHA, DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, ... | | 著者 | Petzold, G, Fischer, E.S, Thoma, N.H. | | 登録日 | 2015-12-09 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural Basis of Lenalidomide-Induced Ck1Alpha Degradation by the Crl4(Crbn) Ubiquitin Ligase.
Nature, 532, 2016
|
|
5TTE
 
 | | Crystal Structure of an RBR E3 ubiquitin ligase in complex with an E2-Ub thioester intermediate mimic | | 分子名称: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | 著者 | Yuan, L, Lv, Z, Olsen, S.K. | | 登録日 | 2016-11-03 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.501 Å) | | 主引用文献 | Structural insights into the mechanism and E2 specificity of the RBR E3 ubiquitin ligase HHARI.
Nat Commun, 8, 2017
|
|
3BI7
 
 | | Crystal structure of the SRA domain of E3 ubiquitin-protein ligase UHRF1 | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION, ... | | 著者 | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2007-11-30 | | 公開日 | 2007-12-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1.
Nature, 455, 2008
|
|
3CLZ
 
 | | The set and ring associated (SRA) domain of UHRF1 bound to methylated DNA | | 分子名称: | 5'-D(*DCP*DCP*DCP*DTP*DGP*DCP*DGP*DGP*DGP*DCP*DCP*DC)-3', 5'-D(*DGP*DGP*DGP*DCP*DCP*(5CM)P*DGP*DCP*DAP*DGP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1 | | 著者 | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2008-03-20 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1.
Nature, 455, 2008
|
|
2RNO
 
 | |
8QFC
 
 | | UFL1 E3 ligase bound 60S ribosome | | 分子名称: | 60S ribosomal protein L10a, CDK5 regulatory subunit-associated protein 3, DDRGK domain-containing protein 1, ... | | 著者 | Makhlouf, L, Zeqiraj, E, Kulathu, Y. | | 登録日 | 2023-09-04 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
8ENI
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | 分子名称: | 3-[4-(5-fluoro-4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)butyl]-5-methyl-1,3-benzoxazol-2(3H)-one, Bifunctional ligase/repressor BirA | | 著者 | Wilce, M.C.J, Cini, D.A. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
6SJ7
 
 | |
5H7S
 
 | |
6R7N
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | 登録日 | 2019-03-29 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
8ROY
 
 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | 分子名称: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | 著者 | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | 登録日 | 2024-01-12 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
8ROX
 
 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12 | | 分子名称: | 5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-~{N},~{N},3-trimethyl-furan-2-carboxamide;ethane, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | 著者 | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | 登録日 | 2024-01-12 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
4NDF
 
 | | Human Aprataxin (Aptx) bound to RNA-DNA, AMP, and Zn - product complex | | 分子名称: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | 著者 | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | 登録日 | 2013-10-26 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.944 Å) | | 主引用文献 | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDI
 
 | | Human Aprataxin (Aptx) AOA1 variant K197Q bound to RNA-DNA, AMP, and Zn - product complex | | 分子名称: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | 著者 | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | 登録日 | 2013-10-26 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDH
 
 | | Human Aprataxin (Aptx) bound to DNA, AMP, and Zn - product complex | | 分子名称: | 5'-D(P*GP*TP*TP*CP*TP*AP*GP*AP*AP*C)-3', ADENOSINE MONOPHOSPHATE, Aprataxin, ... | | 著者 | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | 登録日 | 2013-10-26 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.848 Å) | | 主引用文献 | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDG
 
 | | Human Aprataxin (Aptx) bound to RNA-DNA and Zn - adenosine vanadate transition state mimic complex | | 分子名称: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', Aprataxin, ... | | 著者 | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | 登録日 | 2013-10-26 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.541 Å) | | 主引用文献 | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
8D4X
 
 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a dimeric form | | 分子名称: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | 著者 | Wang, F, He, Q, Lin, G, Li, H. | | 登録日 | 2022-06-02 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
8E0Q
 
 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a C2 symmetric dimeric form | | 分子名称: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | 著者 | Wang, F, He, Q, Lin, G, Li, H. | | 登録日 | 2022-08-09 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
8EWI
 
 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a tetrameric form | | 分子名称: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | 著者 | Wang, F, He, Q, Lin, G, Li, H. | | 登録日 | 2022-10-23 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
7K04
 
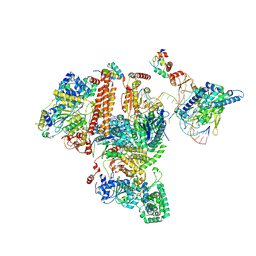 | | Structure of TFIIH/Rad4-Rad23-Rad33/DNA in DNA opening | | 分子名称: | CALCIUM ION, DNA repair helicase RAD25, DNA repair helicase RAD3, ... | | 著者 | van Eeuwen, T, Min, J.H, Murakami, K. | | 登録日 | 2020-09-03 | | 公開日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (9.25 Å) | | 主引用文献 | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
2HVQ
 
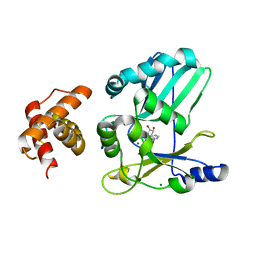 | | Structure of Adenylated full-length T4 RNA Ligase 2 | | 分子名称: | Hypothetical 37.6 kDa protein in Gp24-hoc intergenic region, MAGNESIUM ION | | 著者 | Nandakumar, J, Lima, C.D. | | 登録日 | 2006-07-30 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | 分子名称: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | 登録日 | 2023-07-11 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8B9B
 
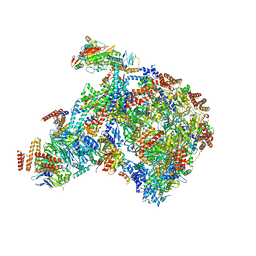 | |
8B9A
 
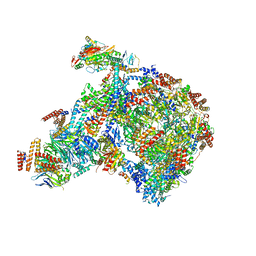 | |
