9IIR
 
 | | human alpha 7 nicotinic acetylcholine receptor in complex with GAT107 and calcium (desensitized state) | | 分子名称: | (3aR,4S,9bS)-4-(4-bromophenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Liu, S, Zheng, Y, Tian, C. | | 登録日 | 2024-06-21 | | 公開日 | 2025-04-23 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural basis for allosteric agonism of human alpha 7 nicotinic acetylcholine receptors.
Cell Discov, 11, 2025
|
|
9FQP
 
 | | EGFR Exon20 insertion mutant NPG bound with Compound 23 | | 分子名称: | (7~{S})-3-[(3-chloranyl-2-methoxy-phenyl)amino]-2-(3-fluoranylpyridin-4-yl)-7-(2-methoxyethyl)-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, Epidermal growth factor receptor | | 著者 | Hilbert, B.J, Brooijmans, N, Milgram, B.C, Pagliarini, R.A. | | 登録日 | 2024-06-17 | | 公開日 | 2025-01-29 | | 最終更新日 | 2025-02-26 | | 実験手法 | X-RAY DIFFRACTION (2.495 Å) | | 主引用文献 | Discovery of STX-721, a Covalent, Potent, and Highly Mutant-Selective EGFR/HER2 Exon20 Insertion Inhibitor for the Treatment of Non-Small Cell Lung Cancer.
J.Med.Chem., 68, 2025
|
|
8OQA
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with GABA and picrotoxin | | 分子名称: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | 登録日 | 2023-04-11 | | 公開日 | 2023-08-30 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OQ8
 
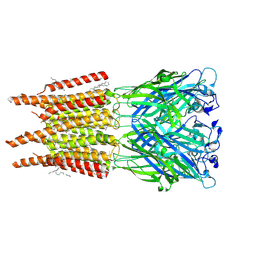 | | CryoEM structure of human rho1 GABAA receptor in complex with pore blocker picrotoxin | | 分子名称: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, DECANE, ... | | 著者 | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | 登録日 | 2023-04-11 | | 公開日 | 2023-08-30 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
1ALF
 
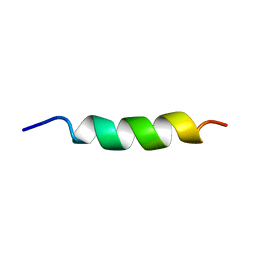 | |
6CRH
 
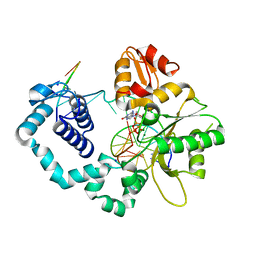 | |
3E7S
 
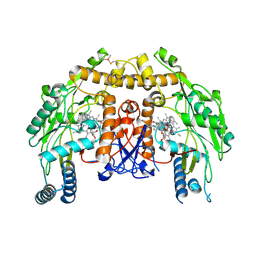 | | Structure of bovine eNOS oxygenase domain with inhibitor AR-C95791 | | 分子名称: | ETHYL 4-[(4-METHYLPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, HEME C, Nitric oxide synthase, ... | | 著者 | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2008-08-18 | | 公開日 | 2008-10-07 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
6Y9P
 
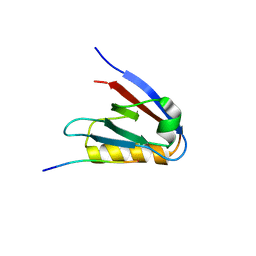 | | Crystal structure of Whirlin PDZ3_C-ter in complex with Harmonin a1 C-terminal PDZ binding motif peptide | | 分子名称: | Harmonin a1, Whirlin | | 著者 | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | 登録日 | 2020-03-10 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.169 Å) | | 主引用文献 | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
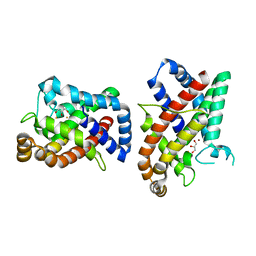
8CQM
 
 | |
6GH0
 
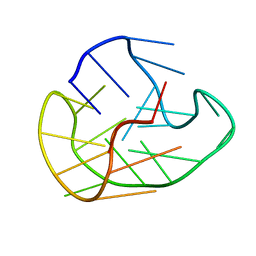 | |
6VHS
 
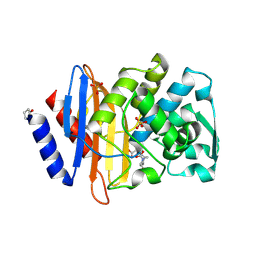 | | Crystal structure of CTX-M-14 in complex with beta-lactamase inhibitor ETX1317 | | 分子名称: | (2R)-({[(3R,6S)-6-carbamoyl-1-formyl-4-methyl-1,2,3,6-tetrahydropyridin-3-yl]amino}oxy)(fluoro)acetic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Sacco, M.D, Chen, Y. | | 登録日 | 2020-01-10 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Discovery of an Orally Available Diazabicyclooctane Inhibitor (ETX0282) of Class A, C, and D Serine beta-Lactamases.
J.Med.Chem., 63, 2020
|
|
1A2E
 
 | |
6H08
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with a His175Me-His proximal ligand substitution | | 分子名称: | COBALT (II) ION, Cytochrome c peroxidase, mitochondrial, ... | | 著者 | Ortmayer, M, Levy, C, Green, A.P. | | 登録日 | 2018-07-06 | | 公開日 | 2020-02-12 | | 最終更新日 | 2025-04-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Rewiring the "Push-Pull" Catalytic Machinery of a Heme Enzyme Using an Expanded Genetic Code.
Acs Catalysis, 10, 2020
|
|
7RFB
 
 | |
7RFC
 
 | |
4M3V
 
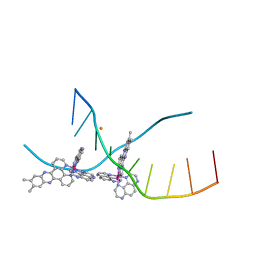 | | X-ray crystal structure of the ruthenium complex [Ru(Tap)2(dppz-{Me2})]2+ bound to d(TCGGTACCGA) | | 分子名称: | (11,12-dimethyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium(2+), BARIUM ION, DNA decamer sequence | | 著者 | Niyazi, H, Teixeira, S, Mitchell, E, Forsyth, T, Cardin, C. | | 登録日 | 2013-08-06 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | X-ray crystal structure of the ruthenium complex [Ru(Tap)2(dppz-{Me2})]2+ bound to d(TCGGTACCGA)
To be Published
|
|
8DHZ
 
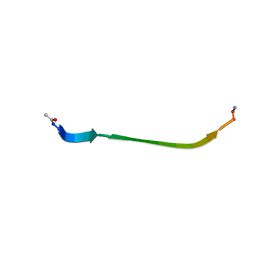 | | NMR Structure of Ac-hGal(17-30)NH2, an N-terminally acetylated fragment of the C-terminus of human galanin | | 分子名称: | Galanin | | 著者 | Kraichely, K.N, Hendy, C.M, Parnham, S, Giuliano, M.W. | | 登録日 | 2022-06-28 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-10-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The neuropeptide galanin adopts an irregular secondary structure.
Biochem.Biophys.Res.Commun., 626, 2022
|
|
8T9A
 
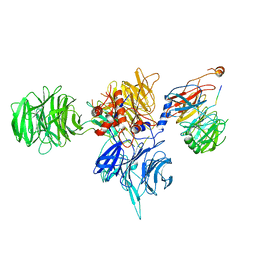 | | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3 | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, Melanoma-associated antigen 3 | | 著者 | Duda, D, Righetto, G, Li, Y, Loppnau, P, Seitova, A, Santhakumar, V, Halabelian, L, Yin, Y. | | 登録日 | 2023-06-23 | | 公開日 | 2024-04-10 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Probing the CRL4 DCAF12 interactions with MAGEA3 and CCT5 di-Glu C-terminal degrons.
Pnas Nexus, 3, 2024
|
|
6CV9
 
 | |
6GJH
 
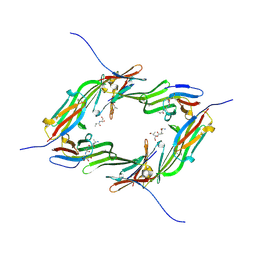 | |
7XQF
 
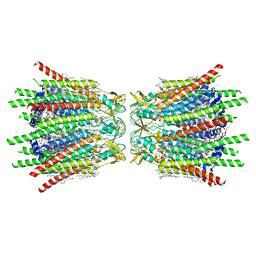 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | 著者 | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | 登録日 | 2022-05-07 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQD
 
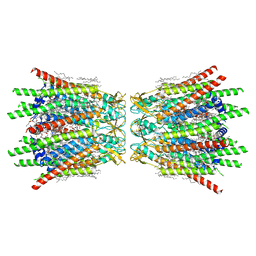 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs (C1 symmetry) | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | 著者 | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | 登録日 | 2022-05-07 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
8OGS
 
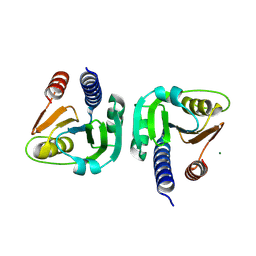 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry B08 | | 分子名称: | Cyclic di-AMP synthase CdaA, MAGNESIUM ION, ethyl 1,3-dihydro-2H-pyrrolo[3,4-c]pyridine-2-carboxylate | | 著者 | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | 登録日 | 2023-03-20 | | 公開日 | 2024-03-27 | | 最終更新日 | 2025-05-14 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
4O79
 
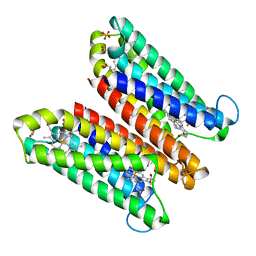 | | Crystal Structure of Ascorbate-bound Cytochrome b561, crystal soaked in 1 M L-ascorbate for 10 minutes | | 分子名称: | ASCORBIC ACID, PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, ... | | 著者 | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | 登録日 | 2013-12-24 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5HHH
 
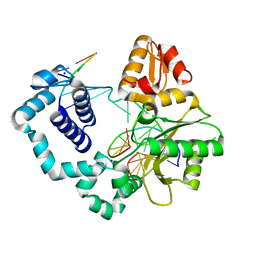 | | Structure of human DNA polymerase beta Host-Guest complexed with the control G for N7-CBZ-platination | | 分子名称: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*AP*GP*GP*AP*GP*CP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | 著者 | Koag, M.-C, Lee, S. | | 登録日 | 2016-01-11 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.363 Å) | | 主引用文献 | Synthesis, structure, and biological evaluation of a platinum-carbazole conjugate.
Chem Biol Drug Des, 91, 2018
|
|
