1D1N
 
 | | SOLUTION STRUCTURE OF THE FMET-TRNAFMET BINDING DOMAIN OF BECILLUS STEAROTHERMOPHILLUS TRANSLATION INITIATION FACTOR IF2 | | Descriptor: | INITIATION FACTOR 2 | | Authors: | Meunier, S, Spurio, S, Czisch, M, Wechselberger, R, Gueunneugues, M. | | Deposit date: | 1999-09-20 | | Release date: | 2000-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the fMet-tRNA(fMet)-binding domain of B. stearothermophilus initiation factor IF2.
EMBO J., 19, 2000
|
|
3E1Y
 
 | | Crystal structure of human eRF1/eRF3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, Eukaryotic peptide chain release factor subunit 1 | | Authors: | Cheng, Z, Lim, M, Kong, C, Song, H. | | Deposit date: | 2008-08-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural insights into eRF3 and stop codon recognition by eRF1
Genes Dev., 23, 2009
|
|
5VH6
 
 | | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis. | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis.
To Be Published
|
|
5JUO
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure I (fully rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
7QTT
 
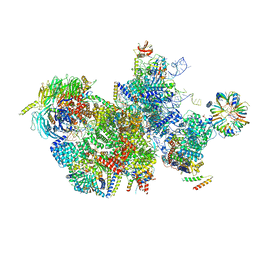 | |
6D9J
 
 | | Mammalian 80S ribosome with a double translocated CrPV-IRES, P-sitetRNA and eRF1. | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Pisareva, V.P, Pisarev, A.V, Fernandez, I.S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-06-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES.
Elife, 7, 2018
|
|
2FX3
 
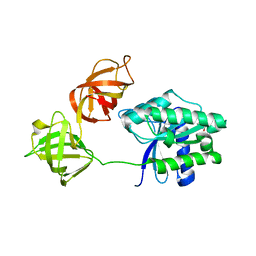 | | Crystal Structure Determination of E. coli Elongation Factor, Tu using a Twinned Data Set | | Descriptor: | Elongation factor Tu, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Heffron, S.E, Moeller, R, Jurnak, F. | | Deposit date: | 2006-02-03 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Solving the structure of Escherichia coli elongation factor Tu using a twinned data set.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4FN5
 
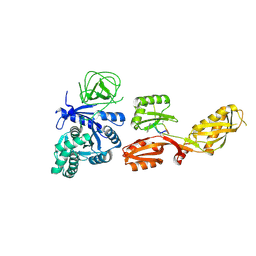 | |
1N0V
 
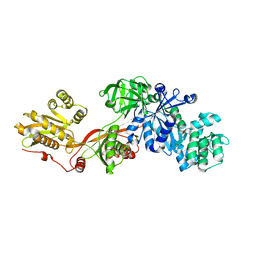 | | Crystal structure of elongation factor 2 | | Descriptor: | Elongation factor 2 | | Authors: | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat.Struct.Biol., 10, 2003
|
|
8EWH
 
 | | Salmonella typhimurium GTPase BIPA | | Descriptor: | 50S ribosomal subunit assembly factor BipA, SODIUM ION | | Authors: | Brown, R.S, deLivron, M.A, Robinson, V.L. | | Deposit date: | 2022-10-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic and Biochemical Characterization of the GTPase and Ribosome Binding Properties of Salmonella typhimuirum BipA
J Biomol Struct Dyn., 24:6, 2007
|
|
5FG3
 
 | | Crystal structure of GDP-bound aIF5B from Aeropyrum pernix | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Probable translation initiation factor IF-2 | | Authors: | Murakami, R, Miyoshi, T, Uchiumi, T, Ito, K. | | Deposit date: | 2015-12-20 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of translation initiation factor 5B from the crenarchaeon Aeropyrum pernix.
Proteins, 84, 2016
|
|
3MMP
 
 | | Structure of the Qb replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Elongation factor Tu 2, Elongation factor Ts, ... | | Authors: | Kidmose, R.T, Vasiliev, N.N, Chetverin, A.B, Knudsen, C.R, Andersen, G.R. | | Deposit date: | 2010-04-20 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8RC0
 
 | | Structure of the human 20S U5 snRNP | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6K72
 
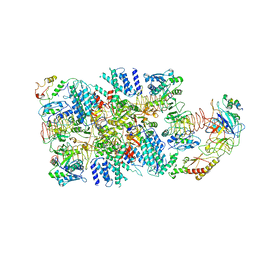 | | eIF2(aP) - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Kashiwagi, K, Yokoyama, T, Ito, T. | | Deposit date: | 2019-06-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
6QX9
 
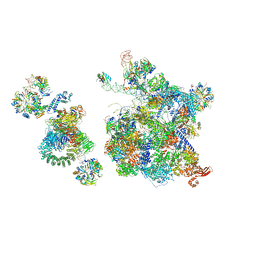 | | Structure of a human fully-assembled precatalytic spliceosome (pre-B complex). | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, AdML pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Charenton, C, Wilkinson, M.E, Nagai, K. | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Mechanism of 5' splice site transfer for human spliceosome activation.
Science, 364, 2019
|
|
7LS1
 
 | | 80S ribosome from mouse bound to eEF2 (Class II) | | Descriptor: | 28S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Loerch, S, Smith, P.R, Kunder, N, Stanowick, A.D, Lou, T.-F, Campbell, Z.T. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance.
Nat Commun, 12, 2021
|
|
7LS2
 
 | | 80S ribosome from mouse bound to eEF2 (Class I) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loerch, S, Smith, P.R, Kunder, N, Stanowick, A.D, Lou, T.-F, Campbell, Z.T. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance.
Nat Commun, 12, 2021
|
|
2RDO
 
 | | 50S subunit with EF-G(GDPNP) and RRF bound | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Gao, N, Zavialov, A.V, Ehrenberg, M, Frank, J. | | Deposit date: | 2007-09-24 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits.
J.Mol.Biol., 374, 2007
|
|
6Z6M
 
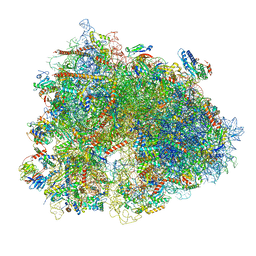 | | Cryo-EM structure of human 80S ribosomes bound to EBP1, eEF2 and SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
6K71
 
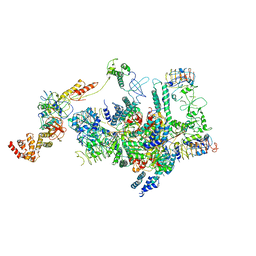 | | eIF2 - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Kashiwagi, K, Yokoyama, T, Ito, T. | | Deposit date: | 2019-06-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
6Z6N
 
 | | Cryo-EM structure of human EBP1-80S ribosomes (focus on EBP1) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
8RM5
 
 | | Cryo-EM structure of the cross-exon pre-B+5'ssLNG+ATPyS complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'SS oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2024-01-05 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
6MIJ
 
 | | Crystal structure of EF-Tu from Acinetobacter baumannii in complex with Mg2+ and GDP | | Descriptor: | Elongation factor Tu, FORMIC ACID, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-19 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | To be published
To Be Published
|
|
6NOT
 
 | |
6RA9
 
 | | Novel structural features and post-translational modifications in eukaryotic elongation factor 1A2 from Oryctolagus cuniculus | | Descriptor: | Elongation factor 1-alpha 2, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Carriles, A.A, Hermoso, J, Gago, F. | | Deposit date: | 2019-04-05 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Cues for Understanding eEF1A2 Moonlighting.
Chembiochem, 22, 2021
|
|
