5RTS
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000159004 | | Descriptor: | 5-phenylpyridine-3-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
6E73
 
 | | Structure of Human Transthyretin Val30Met Mutant in Complex with Diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, ACETATE ION, Transthyretin | | Authors: | Chung, K, Saelices, L, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structural Variants of Transthyretin
to be published
|
|
7R61
 
 | | BTK in complex with 25A | | Descriptor: | 5-{(3S)-1-[(Z)-iminomethyl]piperidin-3-yl}-2-(4-phenoxyphenoxy)pyridine-3-carboxamide, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A. | | Deposit date: | 2021-06-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of Covalent Bruton's Tyrosine Kinase Inhibitors with Decreased CYP2C8 Inhibitory Activity.
Chemmedchem, 16, 2021
|
|
4ZTM
 
 | | Irak4-inhibitor co-structure | | Descriptor: | 5-(1,3-benzothiazol-2-yl)-2-(cyclopropylamino)-6-{[(1R,2S,3R,4R)-2,3-dihydroxy-4-(hydroxymethyl)cyclopentyl]amino}pyrimidin-4(3H)-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery and Structure Enabled Synthesis of 2,6-Diaminopyrimidin-4-one IRAK4 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5AI6
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 5-BROMOQUINOLINE, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-02-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
8C4F
 
 | | Small molecule amidine soak in 14-3-3/ERa (AZ037) | | Descriptor: | 14-3-3 protein sigma, 5-(cyclohexylamino)-4-phenyl-thiophene-2-carboximidamide, ERalpha peptide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2023-01-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HZ9
 
 | | Crystal structure of AtHPPD-Y181136 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-methyl-6-[(2-methyl-3-oxidanylidene-1H-pyrazol-4-yl)carbonyl]-3-propan-2-yl-1,2,3-benzotriazin-4-one, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-08 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal structure of AtHPPD-Y181136 complex
To Be Published
|
|
6E70
 
 | | Structure of Wild Type Human Transthyretin in Complex with Diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, CALCIUM ION, Transthyretin | | Authors: | Chung, K, Saelices, L, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
6E78
 
 | | Structure of Human Transthyretin Asp38Ala Mutant in Complex with Diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, Transthyretin | | Authors: | Chung, K, Saelices, L, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
6E97
 
 | |
4ZTN
 
 | | Irak4-inhibitor co-structure | | Descriptor: | 5-(1,3-benzothiazol-2-yl)-2-(morpholin-4-yl)-6-[(3R)-piperidin-3-ylamino]pyrimidin-4(3H)-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and Structure Enabled Synthesis of 2,6-Diaminopyrimidin-4-one IRAK4 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
7YCQ
 
 | |
5JWC
 
 | | Structure of NDH2 from plasmodium falciparum in complex with RYL-552 | | Descriptor: | 5-fluoro-3-methyl-2-{4-[4-(trifluoromethoxy)benzyl]phenyl}quinolin-4(1H)-one, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Yu, Y. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
6MWJ
 
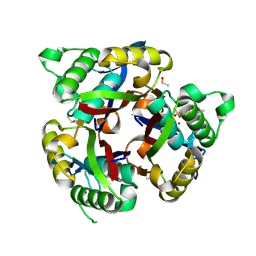 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (IspF) Burkholderia pseudomallei in complex with ligand HGN-0863 | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, DIMETHYL SULFOXIDE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Analysis of Burkholderia pseudomallei IspF in complex with sulfapyridine, sulfamonomethoxine, ethoxzolamide and acetazolamide
Acta Crystallogr.,Sect.F, 81, 2025
|
|
7Z1G
 
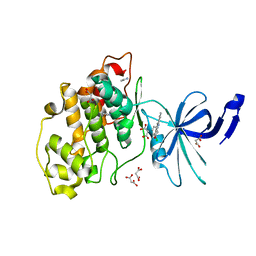 | | Crystal structure of nonphosphorylated (Tyr216) GSK3b in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Glycogen synthase kinase-3 beta, IMIDAZOLE, ... | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-02-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of nonphosphorylated (Tyr216) GSK3b kinase in complex with CX-4945
To Be Published
|
|
7Z1F
 
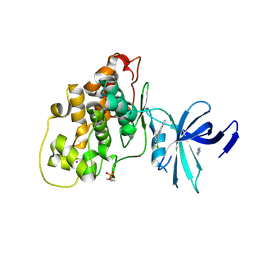 | | Crystal structure of GSK3b in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Glycogen synthase kinase-3 beta, IMIDAZOLE, ... | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-02-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of phosphorylated (Tyr216) GSK3b kinase in complex with CX-4945
To Be Published
|
|
6KHF
 
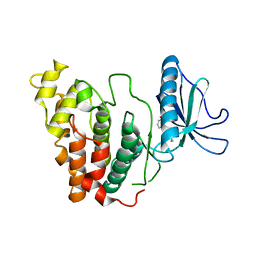 | | Crystal structure of CLK3 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK3 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
6KCC
 
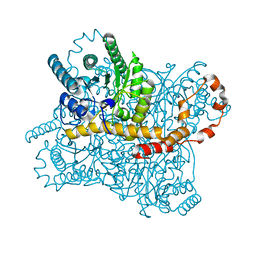 | |
6KCA
 
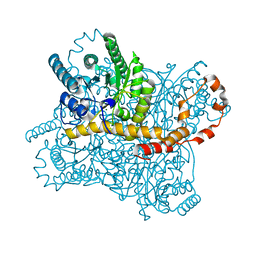 | |
6KHD
 
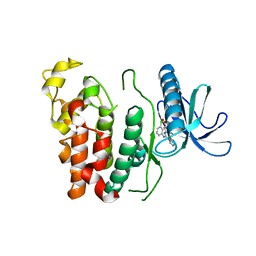 | | Crystal structure of CLK1 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK1 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
1IE8
 
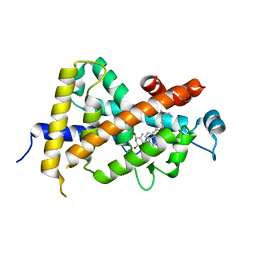 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060 | | Descriptor: | 5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4OXY
 
 | | Substrate-binding loop movement with inhibitor PT10 in the tetrameric Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 5-hexyl-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Sullivan, T.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4WR8
 
 | | Macrophage Migration Inhibitory Factor in complex with a biaryltriazole inhibitor (3b-180) | | Descriptor: | 4-[4-(quinolin-2-yl)-1H-1,2,3-triazol-1-yl]phenol, Macrophage migration inhibitory factor, SODIUM ION, ... | | Authors: | Robertson, M.J, Baxter, R.H.G, Jorgensen, W.L. | | Deposit date: | 2014-10-23 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, and protein crystallography of biaryltriazoles as potent tautomerase inhibitors of macrophage migration inhibitory factor.
J.Am.Chem.Soc., 137, 2015
|
|
8XIA
 
 | |
4QDW
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Ni2+ ions and linear L-arabinose | | Descriptor: | L-arabinose, NICKEL (II) ION, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
