6HYI
 
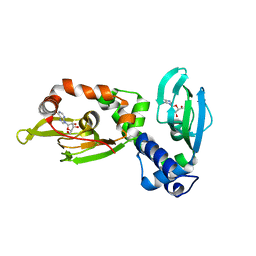 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma cruzi at 1.4 A resolution in complex with inosine | | Descriptor: | INOSINE, Protein kinase A regulatory subunit | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.39944851 Å) | | Cite: | Purine nucleosides replace cAMP in allosteric regulation of PKA in trypanosomatid pathogens.
Elife, 12, 2024
|
|
4K5A
 
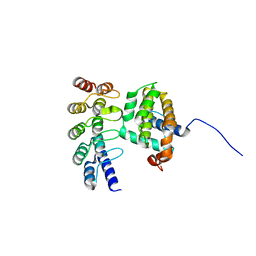 | | Co-crystallization with conformation-specific designed ankyrin repeat proteins explains the conformational flexibility of BCL-W | | Descriptor: | Bcl-2-like protein 2, Designed Ankyrin Repeat Protein 013_D12 | | Authors: | Schilling, J, Schoeppe, J, Sauer, E, Plueckthun, A. | | Deposit date: | 2013-04-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-Crystallization with Conformation-Specific Designed Ankyrin Repeat Proteins Explains the Conformational Flexibility of BCL-W
J.Mol.Biol., 426, 2014
|
|
8DN5
 
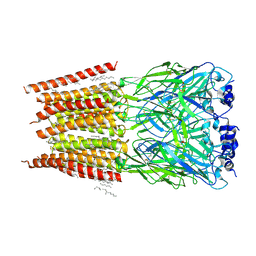 | |
8DN4
 
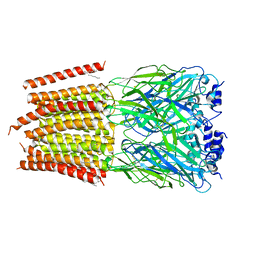 | | Cryo-EM structure of human Glycine Receptor alpha-1 beta heteromer, glycine-bound state3(desensitized state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-1, Glycine receptor subunit beta,Green fluorescent protein,Glycine receptor beta, ... | | Authors: | Liu, X, Wang, W. | | Deposit date: | 2022-07-10 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric gating of a human hetero-pentameric glycine receptor.
Nat Commun, 14, 2023
|
|
8DN3
 
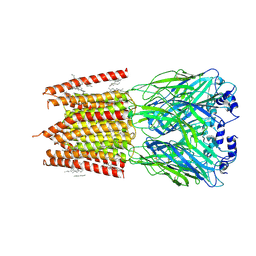 | |
4YCV
 
 | |
8E99
 
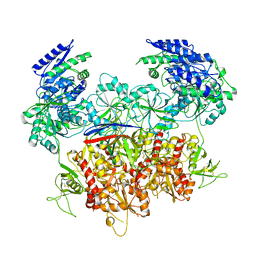 | | Human GluN1a-GluN2A-GluN2C triheteromeric NMDA receptor in complex with Nb-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E98
 
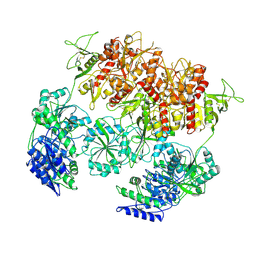 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in nanodisc - intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
3MMF
 
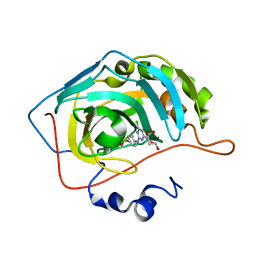 | | Crystal structure of human carbonic anhydrase II in complex with a 1,3,5-triazine-substituted benzenesulfonamide inhibitor | | Descriptor: | 4-({4-chloro-6-[(2-hydroxyethyl)amino]-1,3,5-triazin-2-yl}amino)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, Mckenna, R. | | Deposit date: | 2010-04-19 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sulfonamides incorporating 1,3,5-triazine moieties selectively and potently inhibit carbonic anhydrase transmembrane isoforms IX, XII and XIV over cytosolic isoforms I and II: Solution and X-ray crystallographic studies.
Bioorg.Med.Chem., 19, 2011
|
|
3FCU
 
 | | Structure of headpiece of integrin aIIBb3 in open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
6GTI
 
 | | Achromobacter cycloclastes copper nitrite reductase at pH 5.0 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Halsted, T.P, Eady, R.R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
6GTK
 
 | | Achromobacter cycloclastes copper nitrite reductase at pH 5.5 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Halsted, T.P, Eady, R.R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
7TTP
 
 | | P450 (OxyA) from kistamicin biosynthesis, mixed heme conformation | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 hydroxylase | | Authors: | Greule, A, Izore, T, Cryle, M.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade is Highly Sensitive to Oxidative Damage.
Front Chem, 10, 2022
|
|
7TTA
 
 | | P450 (OxyA) from kistamicin biosynthesis, mixed heme conformation, attenuated beam | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase | | Authors: | Greule, A, Izore, T, Cryle, M.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade is Highly Sensitive to Oxidative Damage.
Front Chem, 10, 2022
|
|
7TTQ
 
 | | P450 (OxyA) from kistamicin biosynthesis, imidazole complex | | Descriptor: | GLYCEROL, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Greule, A, Izore, T, Cryle, M.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade is Highly Sensitive to Oxidative Damage.
Front Chem, 10, 2022
|
|
6Q6G
 
 | | Cryo-EM structure of the APC/C-Cdc20-Cdk2-cyclinA2-Cks2 complex, the D1 box class | | Descriptor: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Zhang, S, Barford, D. | | Deposit date: | 2018-12-11 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C.
Nat Commun, 10, 2019
|
|
7TTO
 
 | |
7TTB
 
 | | P450 (OxyA) from kistamicin biosynthesis, Y99F mutant | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase | | Authors: | Greule, A, Izore, T, Cryle, M.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.801592 Å) | | Cite: | The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade is Highly Sensitive to Oxidative Damage.
Front Chem, 10, 2022
|
|
6DM1
 
 | | Open state GluA2 in complex with STZ and blocked by NASPM, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
3IBD
 
 | | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Gay, S.C, Sun, L, Talakad, J.C, Shah, M.B, Stout, D.C, Halpert, J.R. | | Deposit date: | 2009-07-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-A resolution.
Mol.Pharmacol., 77, 2010
|
|
3FN1
 
 | | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. | | Descriptor: | NEDD8-activating enzyme E1 catalytic subunit, NEDD8-conjugating enzyme UBE2F | | Authors: | Huang, D.T, Ayrault, O, Hunt, H.W, Taherbhoy, A.M, Duda, D.M, Scott, D.C, Borg, L.A, Neale, G, Murray, P.J, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification
Mol.Cell, 33, 2009
|
|
4B3W
 
 | | Crystal structure of human cytoglobin H(E7)Q mutant | | Descriptor: | ACETATE ION, CYANIDE ION, CYTOGLOBIN, ... | | Authors: | Gabba, M, Abbruzzetti, S, Spyrakis, F, Forti, F, Bruno, S, Mozzarelli, A, Luque, F.J, Viappiani, C, Cozzini, P, Nardini, M, Germani, F, Bolognesi, M, Moens, L, Dewilde, S. | | Deposit date: | 2012-07-26 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Co Rebinding Kinetics and Molecular Dynamics Simulations Highlight Dynamic Regulation of Internal Cavities in Human Cytoglobin.
Plos One, 8, 2013
|
|
1W4Y
 
 | | Ferrous horseradish peroxidase C1A in complex with carbon monoxide | | Descriptor: | CALCIUM ION, CARBON MONOXIDE, HORSERADISH PEROXIDASE C1A, ... | | Authors: | Carlsson, G.H, Nicholls, P, Svistunenko, D, Berglund, G.I, Hajdu, J. | | Deposit date: | 2004-08-03 | | Release date: | 2005-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complexes of Horseradish Peroxidase with Formate, Acetate, and Carbon Monoxide
Biochemistry, 44, 2005
|
|
6GTN
 
 | | Achromobacter cycloclastes copper nitrite reductase at pH 6.5 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Halsted, T.P, Eady, R.R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
4WR7
 
 | | Crystal structure of human carbonic anhydrase isozyme I with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide. | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-(propylsulfanyl)benzenesulfonamide, ACETATE ION, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-10-23 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
