6R1M
 
 | | Crystal structure of E. coli seryl-tRNA synthetase complexed to seryl sulfamoyl adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, PHOSPHATE ION, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
8QHK
 
 | | Crystal structure of reduced respiratory Complex I subunits NuoEF from Aquifex aeolicus bound to reduced 3-acetylpyridine adenine dinucleotide | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETYL PYRIDINE ADENINE DINUCLEOTIDE, REDUCED, ... | | Authors: | Wohlwend, D, Friedrich, T, Bucka, S. | | Deposit date: | 2023-09-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of 3-acetylpyridine adenine dinucleotide and ADP-ribose bound to the electron input module of respiratory complex I.
Structure, 32, 2024
|
|
6ZFS
 
 | | Crystal structure of bovine cytochrome bc1 in complex with quinolone inhibitor WDH-1U-4 | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Amporndanai, K, O'Neill, P.M, Hong, W.D, Amewu, R.K, Pidathala, C, Berry, N.G, Biagini, G.A, Leung, S.C, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Targeting the ubiquinol-reduction (Qi) site of the mitochondrial cytochrome bc1 complex for the development of next generation quinolone antimalarials
To Be Published
|
|
5FW0
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(2-(5-((2-methoxyethyl)(methyl)amino)pyridin-3-yl) ethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-(5-((2-methoxyethyl)(methyl)amino)pyridin-3-yl)ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2016-02-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibition by Optimization of the 2-Aminopyridine-Based Scaffold with a Pyridine Linker.
J.Med.Chem., 59, 2016
|
|
7ZFM
 
 | | Engineered Protein Targeting the Zika Viral Envelope Fusion Loop | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Athayde, D, Archer, M, Viana, I.F.T, Adan, W.C.S, Xavier, L.S.S, Lins, R.D. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | In Vitro Neutralisation of Zika Virus by an Engineered Protein Targeting the Viral Envelope Fusion Loop
SSRN, 2022
|
|
6ZFT
 
 | | Crystal structure of bovine cytochrome bc1 in complex with quinolone inhibitor CK-2-68 | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Amporndanai, K, O'Neill, P.M, Hong, W.D, Amewu, R.K, Pidathala, C, Berry, N.G, Biagini, G.A, Leung, S.C, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Targeting the ubiquinol-reduction (Qi) site of the mitochondrial cytochrome bc1 complex for the development of next generation quinolone antimalarials
To Be Published
|
|
7ZOW
 
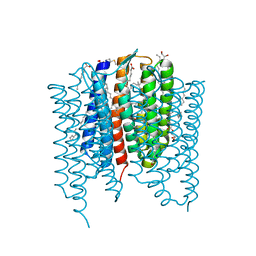 | | Crystal structure of Synechocystis halorhodopsin (SyHR), Cl-pumping mode, O state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Bukhdruker, S, Astashkin, R, Vaganova, S, Gordeliy, V. | | Deposit date: | 2022-04-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into light-driven anion pumping in cyanobacteria.
Nat Commun, 13, 2022
|
|
5G1Q
 
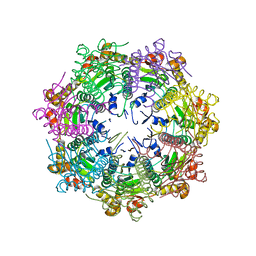 | |
6C7E
 
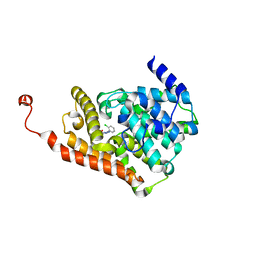 | | Crystal structure of human phosphodiesterase 2A with 1-(2-chlorophenyl)-N,4-dimethyl-[1,2,4]triazolo[4,3-a]quinoxaline-8-carboxamide | | Descriptor: | 1-(2-chlorophenyl)-N,4-dimethyl[1,2,4]triazolo[4,3-a]quinoxaline-8-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2018-01-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Mathematical and Structural Characterization of Strong Nonadditive Structure-Activity Relationship Caused by Protein Conformational Changes.
J. Med. Chem., 61, 2018
|
|
5UQC
 
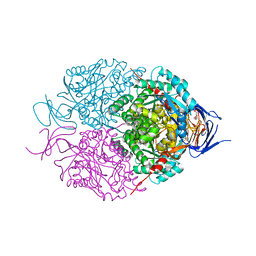 | | Crystal structure of mouse CRMP2 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Dihydropyrimidinase-related protein 2 | | Authors: | Khanna, M, Khanna, R, Perez-Miller, S, Francois-Moutal, L. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A single structurally conserved SUMOylation site in CRMP2 controls NaV1.7 function.
Channels (Austin), 11, 2017
|
|
6Z90
 
 | | Crystal structure of MINDY1 mutant-P138A | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase MINDY-1 | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
6C7W
 
 | | Carbonic anhydrase 2 in complex with [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDRO-2-FURANYL]METHYL SULFAMATE inhibitor | | Descriptor: | Carbonic anhydrase 2, SODIUM ION, ZINC ION, ... | | Authors: | Peat, T.S, Mujumdar, P, Poulsen, S.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Synthesis, structure and bioactivity of primary sulfamate-containing natural products.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
8Q9P
 
 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC5 deacetylase binding motif | | Descriptor: | HDAC5 (histone deacetylase 5) binding motif peptide: TRP-GLY-SER-GLY-GLU-VAL-LYS-LEU-ARG-LEU-GLN-GLU-PHE-LEU-LEU-SER-LYS-SER, MADS box dsDNA fw: AACTATTTATAAGA, MADS box dsNA rev:TCTTATAAATAGTT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
8RHE
 
 | |
6C8B
 
 | |
7MRR
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Leupeptin | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, LEUPEPTIN | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7Q84
 
 | | Crystal structure of human peroxisomal acyl-Co-A oxidase 1a, apo-form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sonani, R.R, Blat, A, Dubin, G. | | Deposit date: | 2021-11-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of apo- and FAD-bound human peroxisomal acyl-CoA oxidase provide mechanistic basis explaining clinical observations.
Int.J.Biol.Macromol., 205, 2022
|
|
6ZFH
 
 | | Structure of human galactokinase in complex with galactose and 2'-(benzo[d]oxazol-2-ylamino)-7',8'-dihydro-1'H-spiro[cyclopentane-1,4'-quinazolin]-5'(6'H)-one | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclopentane]-5-one, Galactokinase, beta-D-galactopyranose | | Authors: | Bezerra, G.A, Mackinnon, S, Zhang, M, Foster, W, Bailey, H, Arrowsmith, C, Edwards, A, Bountra, C, Lai, K, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.439 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6QYC
 
 | |
5URK
 
 | | Crystal structure of human BRR2 in complex with T-3935799 | | Descriptor: | 6-benzyl-3-[3-(benzyloxy)phenyl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(1H,3H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Qin, L, Tjhen, R, Klein, M.G. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
7MF0
 
 | | Co-crystal structure of PERK with inhibitor (R)-2-amino-N-cyclopropyl-5-(4-(2-(3,5-difluorophenyl)-2-hydroxyacetamido)-2-methylphenyl)nicotinamide | | Descriptor: | 2-amino-N-cyclopropyl-5-(4-{[(2R)-2-(3,5-difluorophenyl)-2-hydroxyacetyl]amino}-2-methylphenyl)pyridine-3-carboxamide, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wiens, B, Koszelak-Rosenblum, M, Surman, M.D, Zhu, G, Mulvihill, M.J. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Discovery of 2-amino-3-amido-5-aryl-pyridines as highly potent, orally bioavailable, and efficacious PERK kinase inhibitors.
Bioorg.Med.Chem.Lett., 43, 2021
|
|
6ZE1
 
 | | human NBD1 of CFTR in complex with nanobody G11a | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Garcia-Pino, A, Govaerts, C, Scholl, D, Sigoillot, M. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | A topological switch in CFTR modulates channel activity and sensitivity to unfolding.
Nat.Chem.Biol., 17, 2021
|
|
5G3L
 
 | | ESCHERICHIA COLI HEAT LABILE ENTEROTOXIN TYPE IIB B-PENTAMER COMPLEXED WITH SIALYLATED SUGAR | | Descriptor: | HEAT-LABILE ENTEROTOXIN IIB, B CHAIN, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Zalem, D, Benktander, J, Ribeiro, J.P, Varrot, A, Lebens, M, Imberty, A, Teneberg, S. | | Deposit date: | 2016-04-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Biochemical and Structural Characterization of the Novel Sialic Acid-Binding Site of Escherichia Coli Heat-Labile Enterotoxin Lt-Iib.
Biochem.J., 473, 2016
|
|
6H00
 
 | | Crystal structure of human pyridoxine 5-phophate oxidase, R116Q variant | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Pyridoxine-5'-phosphate oxidase, ... | | Authors: | Mackinnon, S, Wilson, M.P, Shrestha, L, Bezerra, G.A, Newman, J, Fox, N, Sorrell, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Clayton, P.T, Mills, P.B, Yue, W.W. | | Deposit date: | 2018-07-05 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of human pyridoxine 5-phophate oxidase, R116Q variant
To Be Published
|
|
5USF
 
 | |
