7V1V
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium, ligand-free form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
7E2I
 
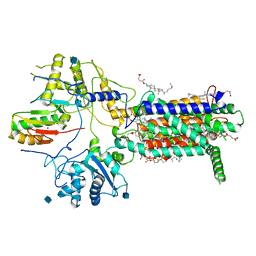 | | Cryo-EM structure of hDisp1NNN-ShhN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1, ... | | Authors: | Li, W, Wang, L, Gong, X. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release.
Nat Commun, 12, 2021
|
|
6P9U
 
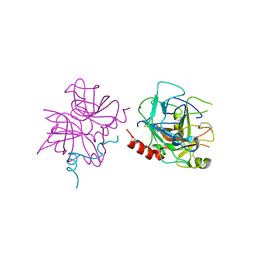 | | Crystal structure of human thrombin mutant W215A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin, ZINC ION | | Authors: | Pelc, L.A, Koester, S.K, Chen, Z, Di Cera, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Residues W215, E217 and E192 control the allosteric E*-E equilibrium of thrombin.
Sci Rep, 9, 2019
|
|
2IQ6
 
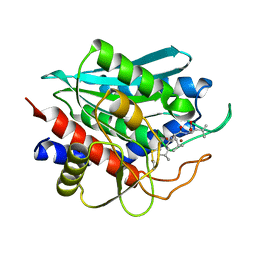 | | Crystal Structure of the Aminopeptidase from Vibrio proteolyticus in Complexation with Leucyl-leucyl-leucine. | | Descriptor: | Bacterial leucyl aminopeptidase, Peptide, (Leucyl-leucyl-leucine), ... | | Authors: | Kumar, A, Narayanan, B, Kim, J.-J.P, Bennett, B. | | Deposit date: | 2006-10-13 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental evidence for a metallohydrolase mechanism in which the nucleophile is not delivered by a metal ion: EPR spectrokinetic and structural studies of aminopeptidase from Vibrio proteolyticus
Biochem.J., 403, 2007
|
|
2EAU
 
 | | Crystal structure of the SR CA2+-ATPASE with bound CPA in the presence of curcumin | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, PHOSPHATIDYLETHANOLAMINE, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2007-02-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1UUD
 
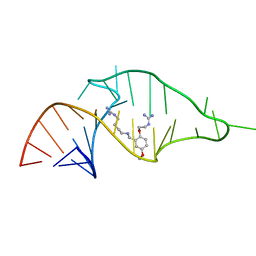 | | NMR structure of a synthetic small molecule, rbt203, bound to HIV-1 TAR RNA | | Descriptor: | N-[2-(2-{[(4-{[AMINO(IMINO)METHYL]AMINO}BUTYL)AMINO]METHYL}-4-METHOXYPHENOXY)ETHYL]GUANIDINE, RNA (5'-(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP *CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C) -3') | | Authors: | Davis, B, Afshar, M, Varani, G, Karn, J, Murchie, A.I.H, Lentzen, G, Drysdale, M.J, Potter, A.J, Bower, J, Aboul-Ela, F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Inhibitors of HIV-1 Tar RNA Through the Stabilisation of Electrostatic "Hot Spots"
J.Mol.Biol., 336, 2004
|
|
6SPP
 
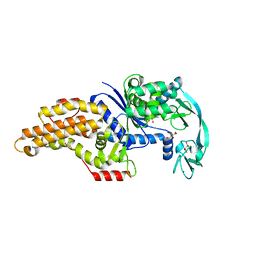 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 | | Descriptor: | CITRIC ACID, GLYCEROL, Methionine--tRNA ligase, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
1DSN
 
 | | D60S N-TERMINAL LOBE HUMAN LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Faber, H.R, Norris, G.E, Baker, E.N. | | Deposit date: | 1995-12-13 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Altered domain closure and iron binding in transferrins: the crystal structure of the Asp60Ser mutant of the amino-terminal half-molecule of human lactoferrin.
J.Mol.Biol., 256, 1996
|
|
6LUO
 
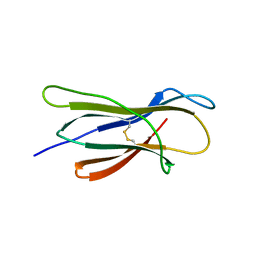 | | Structure of nurse shark beta-2-microglobulin | | Descriptor: | Beta-2-microglobulin | | Authors: | Xia, C, Wu, Y. | | Deposit date: | 2020-01-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between beta 2 -Microglobulin and H Chain Conserved in Evolution.
J Immunol., 2021
|
|
5WBG
 
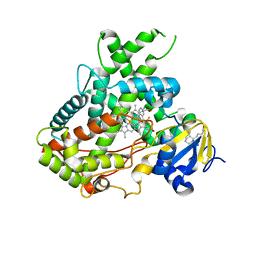 | | Crystal Structure of human Cytochrome P450 2B6 (Y226H/K262R) in complex with an analog of a drug Efavirenz | | Descriptor: | (2R,4S)-6-chloro-4-(cyclopropylethynyl)-2-methyl-4-(trifluoromethyl)-1,4-dihydro-2H-3,1-benzoxazine, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, 5-cyclohexylpentan-1-ol, ... | | Authors: | Shah, M.B, Halpert, J.R. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal Structure of CYP2B6 in Complex with an Efavirenz Analog.
Int J Mol Sci, 19, 2018
|
|
6LUP
 
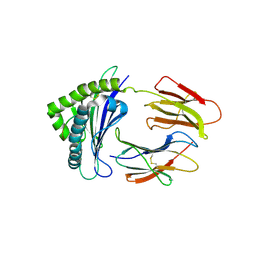 | | Crystal structure of shark MHC CLASS I for 2.3 angstrom | | Descriptor: | Beta-2-microglobulin, MHC class I protein, PHE-ALA-ASN-PHE-PHE-ILE-ARG-GLY-LEU | | Authors: | Wu, Y, Xia, C. | | Deposit date: | 2020-01-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between beta 2 -Microglobulin and H Chain Conserved in Evolution.
J Immunol., 2021
|
|
3RV4
 
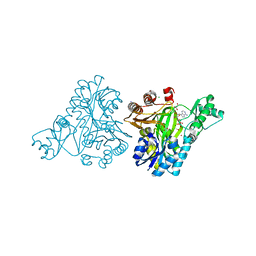 | |
3RUP
 
 | |
7U5P
 
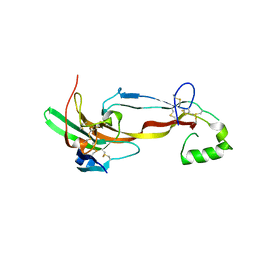 | | CRYSTAL STRUCTURE OF THE ACTIVIN RECEPTOR TYPE-2A LIGAND BINDING DOMAIN IN COMPLEX WITH ACTIVIN-A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2A, Inhibin beta A chain | | Authors: | Chu, K.Y, Malik, A, Thamilselvan, V, Martinez-Hackert, E. | | Deposit date: | 2022-03-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Type II BMP and activin receptors BMPR2 and ACVR2A share a conserved mode of growth factor recognition.
J.Biol.Chem., 298, 2022
|
|
2WHE
 
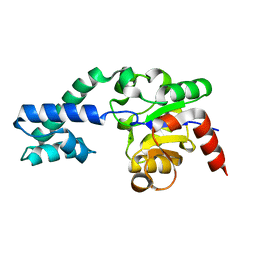 | | Structure of native Beta-Phosphoglucomutase in an open conformation without bound ligands. | | Descriptor: | BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic Details of Near-Transition State Conformers for Enzyme Phosphoryl Transfer Revealed by Mgf-3 Rather Than by Phosphoranes.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1VEB
 
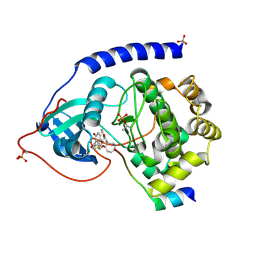 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 5 | | Descriptor: | (3R,4R)-N-{4-[4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-BENZYLOXY]-AZEPAN-3-YL}-ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.-G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
8GJ4
 
 | |
8GKM
 
 | |
8GLC
 
 | |
8GIC
 
 | |
8GJP
 
 | |
4CEV
 
 | | ARGINASE FROM BACILLUS CALDEVELOX, L-ORNITHINE COMPLEX | | Descriptor: | GUANIDINE, L-ornithine, MANGANESE (II) ION, ... | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-15 | | Release date: | 1999-04-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
5EKD
 
 | |
3HKU
 
 | | Human carbonic anhydrase II in complex with topiramate | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3aS,5aR,8aR,8bS)-2,2,7,7-tetramethyltetrahydro-3aH-bis[1,3]dioxolo[4,5-b:4',5'-d]pyran-3a-yl]methyl sulfamate | | Authors: | Paul, B, Poulsen, S.-A, Hofmann, A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S-glycosyl primary sulfonamides--a new structural class for selective inhibition of cancer-associated carbonic anhydrases.
J.Med.Chem., 52, 2009
|
|
3HKT
 
 | | Human carbonic anhydrase II in complex with alpha-D-Glucopyranosyl-(1->4)-1-thio-beta-D-glucopyranosylsulfonamide | | Descriptor: | Carbonic anhydrase 2, ZINC ION, alpha-D-galactopyranose-(1-4)-(1S)-1,5-anhydro-1-sulfamoyl-D-galactitol | | Authors: | Paul, B, Poulsen, S.-A, Hofmann, A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | S-glycosyl primary sulfonamides--a new structural class for selective inhibition of cancer-associated carbonic anhydrases.
J.Med.Chem., 52, 2009
|
|
