6XD3
 
 | | Structure of the human CAK in complex with THZ1 | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Greber, B.J, Perez-Bertoldi, J.M, Lim, K, Iavarone, A.T, Toso, D.B, Nogales, E. | | Deposit date: | 2020-06-09 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The cryoelectron microscopy structure of the human CDK-activating kinase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XEP
 
 | |
6XTX
 
 | | CryoEM structure of human CMG bound to ATPgammaS and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45 homolog, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Rzechorzek, N.J, Pellegrini, L. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | CryoEM structures of human CMG-ATP gamma S-DNA and CMG-AND-1 complexes.
Nucleic Acids Res., 48, 2020
|
|
6DJI
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ522 | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxybenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6XRG
 
 | |
6DJJ
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ532 | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6X65
 
 | | Legionella pneumophila Dot/Icm T4SS | | Descriptor: | DotC, DotD, Inner membrane lipoprotein YiaD, ... | | Authors: | Durie, C.L, Sheedlo, M.J, Chung, J.M, Byrne, B.G, Su, M, Knight, T, Swanson, M.S, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2020-05-27 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural analysis of the Legionella pneumophila Dot/Icm type IV secretion system core complex.
Elife, 9, 2020
|
|
6DLC
 
 | |
6XW5
 
 | | Crystal structure of murine norovirus P domain in complex with Nanobody NB-5820 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Capsid protein, ... | | Authors: | Kilic, T, Sabin, C, Hansman, G. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Nanobody-Mediated Neutralization Reveals an Achilles Heel for Norovirus.
J.Virol., 94, 2020
|
|
6X62
 
 | | Legionella pneumophila Dot T4SS OMC | | Descriptor: | DotC, DotD, Inner membrane lipoprotein YiaD, ... | | Authors: | Durie, C.L, Sheedlo, M.J, Chung, J.M, Byrne, B.G, Su, M, Knight, T, Swanson, M.S, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural analysis of the Legionella pneumophila Dot/Icm type IV secretion system core complex.
Elife, 9, 2020
|
|
6XF8
 
 | | DLP 5 fold | | Descriptor: | Inner capsid protein lambda-1, Inner capsid protein sigma-2, Outer capsid protein mu-1, ... | | Authors: | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
6XHH
 
 | | Far-red absorbing dark state of JSC1_58120g3 with bound 18-1, 18-2 dihydrobiliverdin IXa (DHBV), the native chromophore precursor | | Descriptor: | 1,2-ETHANEDIOL, JSC1_58120g3, mesobiliverdin IX(alpha) | | Authors: | Moreno, M.V, Rockwell, N.C, Fisher, A.J, Lagarias, J.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A far-red cyanobacteriochrome lineage specific for verdins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6DLV
 
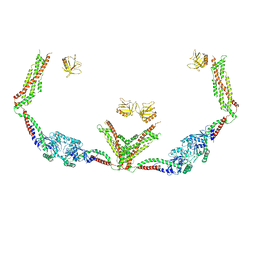 | | Cryo-EM of the GTP-bound human dynamin-1 polymer assembled on the membrane in the super constricted state | | Descriptor: | Dynamin-1 | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
6X9Q
 
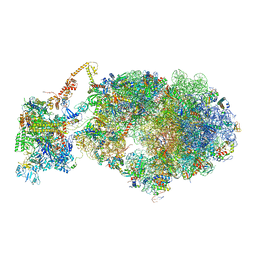 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 27 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
8GYP
 
 | |
8GYT
 
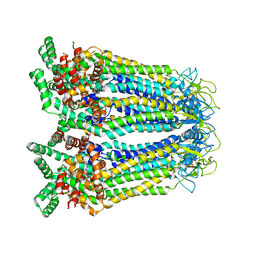 | | Cryo-EM structure of human Pannexin-3 R36S/F40R variant in pre-open state. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pannexin-3 | | Authors: | Cong, Y, Jin, X, Kong, F, Wu, T, Yan, C. | | Deposit date: | 2022-09-23 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Inner channel lipids regulated gating mechanism of human pannexins.
To Be Published
|
|
6DQV
 
 | | Class 2 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6XII
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 24 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-20 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
8T3O
 
 | | Cryo-EM structure of the TUG-891 bound FFA4-Gq complex | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
6DQN
 
 | | Class 1 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6XZA
 
 | | E. coli 70S ribosome in complex with dirithromycin, and deacylated tRNA(iMet) (focused classification). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Pichkur, E.B, Polikanov, Y.S, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2020-02-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Insights into the improved macrolide inhibitory activity from the high-resolution cryo-EM structure of dirithromycin bound to the E. coli 70S ribosome.
Rna, 26, 2020
|
|
6XVT
 
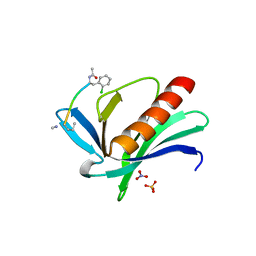 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPPTEDDL-NH2 | | Descriptor: | ACY-SC1-SC2-SC3-SC4-SC5-NME, NITRATE ION, Protein enabled homolog, ... | | Authors: | Barone, M, Le Cong, K, Roske, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XW7
 
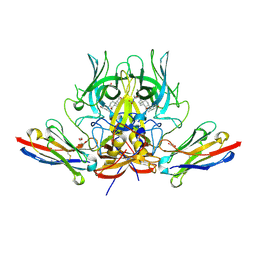 | | Crystal structure of murine norovirus P domain in complex with Nanobody NB-5829 and glycochenodeoxycholate (GCDCA) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, GLYCOCHENODEOXYCHOLIC ACID, ... | | Authors: | Kilic, T, Sabin, C, Hansman, G. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nanobody-Mediated Neutralization Reveals an Achilles Heel for Norovirus.
J.Virol., 94, 2020
|
|
6DW1
 
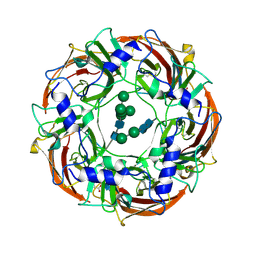 | | Cryo-EM structure of the benzodiazepine-sensitive alpha1beta1gamma2S tri-heteromeric GABAA receptor in complex with GABA (ECD map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1,Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Phulera, S, Zhu, H, Yu, J, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the benzodiazepine-sensitive alpha 1 beta 1 gamma 2S tri-heteromeric GABAAreceptor in complex with GABA.
Elife, 7, 2018
|
|
6XY5
 
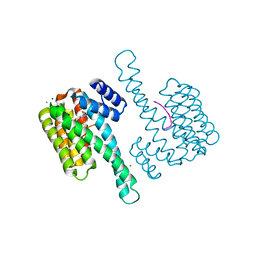 | | Ternary complex of 14-3-3 sigma (C38N), Estrogen Related Receptor gamma (DBD) phosphopeptide, and disulfide PPI stabilizer 5 | | Descriptor: | 14-3-3 protein sigma, 3-bromanyl-~{N}-methyl-~{N}-(2-sulfanylethyl)benzamide, CHLORIDE ION, ... | | Authors: | Somsen, B.A, Ottmann, C. | | Deposit date: | 2020-01-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fluorescence Anisotropy-Based Tethering for Discovery of Protein-Protein Interaction Stabilizers.
Acs Chem.Biol., 15, 2020
|
|
