7D12
 
 | | NMR solution structures of CAG RNA-DB213 binding complex | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*CP*AP*GP*C)-3'), SODIUM ION | | Authors: | Chan, H.Y.E, Guo, P. | | Deposit date: | 2020-09-12 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CAG RNAs induce DNA damage and apoptosis by silencing NUDT16 expression in polyglutamine degeneration.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
6WIV
 
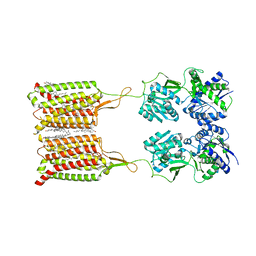 | | Structure of human GABA(B) receptor in an inactive state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-{[(9Z)-octadec-9-enoyl]oxy}propyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Fu, Z, Frangaj, A, Liu, J, Mosyak, L, Shen, T, Slavkovich, V.N, Ray, K.M, Taura, J, Cao, B, Geng, Y, Zuo, H, Kou, Y, Grassucci, R, Chen, S, Liu, Z, Lin, X, Williams, J.P, Rice, W.J, Eng, E.T, Huang, R.K, Soni, R.K, Kloss, B, Yu, Z, Javitch, J.A, Hendrickson, W.A, Slesinger, P.A, Quick, M, Graziano, J, Yu, H, Fiehn, O, Clarke, O.B, Frank, J, Fan, Q.R. | | Deposit date: | 2020-04-10 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human GABABreceptor in an inactive state.
Nature, 584, 2020
|
|
3IWM
 
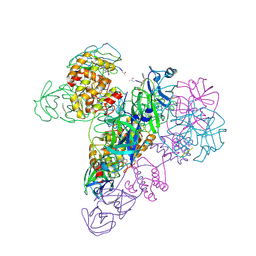 | | The octameric SARS-CoV main protease | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhong, N, Zhang, S, Xue, F, Lou, Z, Rao, Z, Xia, B. | | Deposit date: | 2009-09-02 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease
Protein Cell, 1, 2010
|
|
7EQK
 
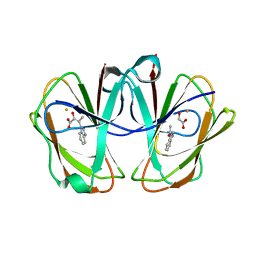 | | Structural and mechanistic studies of a novel non-heme iron epimerase/lyase and its utilization in chemoselective synthesis. | | Descriptor: | (E)-3-(1H-indol-3-yl)-2-oxidanyl-but-2-enoic acid, 1-(1~{H}-indol-3-yl)ethanone, Cupin domain-containing protein, ... | | Authors: | Li, T.L, Li, Y.S, Chen, M.H. | | Deposit date: | 2021-05-03 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04001021 Å) | | Cite: | Structural and Mechanistic Bases for StnK3 and Its Mutant-Mediated Lewis-Acid-Dependent Epimerization and Retro-Aldol Reactions.
Acs Catalysis, 12, 2022
|
|
7XJ3
 
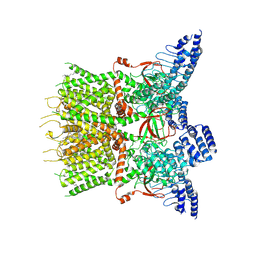 | | Structure of human TRPV3 | | Descriptor: | [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, fusion of transient receptor potential cation channel subfamily V member 3 and 3C-GFP | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ1
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C2 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
3OPV
 
 | | Crystal structure of E. Coli purine nucleoside phosphorylase Arg24Ala mutant | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | Deposit date: | 2010-09-02 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
7D3C
 
 | |
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6W4E
 
 | | NMR-driven structure of KRAS4B-GTP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6UW6
 
 | | Cryo-EM structure of the human TRPV3 K169A mutant determined in lipid nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 3, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Gating of human TRPV3 in a lipid bilayer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7MSM
 
 | | Mtb 70SIC in complex with MtbEttA at Trans_R0 state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7MT2
 
 | | Mtb 70S initiation complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7MSZ
 
 | | Mtb 70SIC in complex with MtbEttA at Trans_R1 state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7MSC
 
 | | Mtb 70SIC in complex with MtbEttA at Pre_R0 state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7MSH
 
 | | Mtb 70SIC in complex with MtbEttA at Pre_R1 state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
3OOH
 
 | | Crystal structure of E. Coli purine nucleoside phosphorylase with PO4 | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
1C8U
 
 | |
6UW4
 
 | | Cryo-EM structure of human TRPV3 determined in lipid nanodisc | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily V member 3, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Gating of human TRPV3 in a lipid bilayer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4GUU
 
 | | Crystal structure of LSD2-NPAC with tranylcypromine | | Descriptor: | Lysine-specific histone demethylase 1B, Putative oxidoreductase GLYR1, ZINC ION, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
7E7O
 
 | | Cryo-EM structure of human ABCA4 in NRPE-bound state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Retinal-specific phospholipid-transporting ATPase ABCA4, ... | | Authors: | Xie, T, Zhang, Z.K, Gong, X. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of substrate recognition and translocation by human ABCA4.
Nat Commun, 12, 2021
|
|
1DNF
 
 | | EFFECTS OF 5-FLUOROURACIL/GUANINE WOBBLE BASE PAIRS IN Z-DNA. MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGFG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(UFP)P*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Saal, D, Frederick, C.A, Aymami, J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-12-12 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effects of 5-fluorouracil/guanine wobble base pairs in Z-DNA: molecular and crystal structure of d(CGCGFG).
Nucleic Acids Res., 17, 1989
|
|
7EW1
 
 | | Cryo-EM structure of siponimod -bound Sphingosine-1-phosphate receptor 5 in complex with Gi protein | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Y, Jia, G.W, Shao, Z.H, Su, Z.M. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
