8ED6
 
 | |
8DPK
 
 | |
8EDA
 
 | | An alternating AT dodecamer benzimidazole (DB1476) complex | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-09-03 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8EC1
 
 | |
1YXA
 
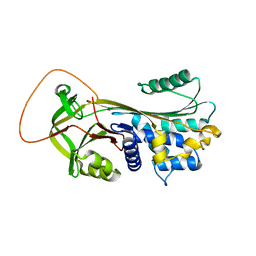 | | Serpina3n, a murine orthologue of human antichymotrypsin | | Descriptor: | serine (or cysteine) proteinase inhibitor, clade A, member 3N | | Authors: | Horvath, A.J, Irving, J.A, Law, R.H, Rossjohn, J, Bottomley, S.P, Quinsey, N.S, Pike, R.N, Coughlin, P.B, Whisstock, J.C. | | Deposit date: | 2005-02-20 | | Release date: | 2005-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins.
J.Biol.Chem., 280, 2005
|
|
2NVO
 
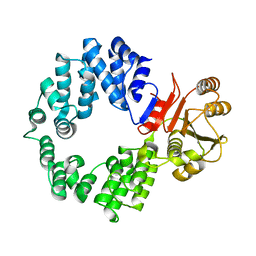 | |
8DWK
 
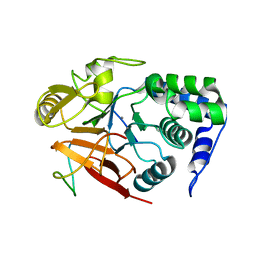 | | Inhibitor-3:PP1 reconstituted complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
6FSH
 
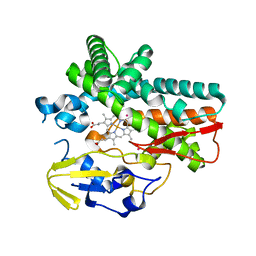 | | Crystal structure of hybrid P450 OxyBtei(BC/FGvan) | | Descriptor: | ACETATE ION, OxyB protein, POTASSIUM ION, ... | | Authors: | Brieke, C, Tarnawski, M, Greule, A, Cryle, M.J. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Investigating Cytochrome P450 specificity during glycopeptide antibiotic biosynthesis through a homologue hybridization approach.
J. Inorg. Biochem., 185, 2018
|
|
8DPB
 
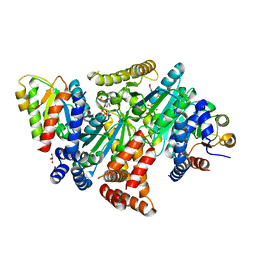 | | MeaB in complex with the cobalamin-binding domain of its target mutase with GMPPCP bound | | Descriptor: | GLYCEROL, MAGNESIUM ION, Methylmalonyl-CoA mutase accessory protein, ... | | Authors: | Vaccaro, F.A, Born, D.A, Drennan, C.L. | | Deposit date: | 2022-07-15 | | Release date: | 2023-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of metallochaperone in complex with the cobalamin-binding domain of its target mutase provides insight into cofactor delivery.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EDB
 
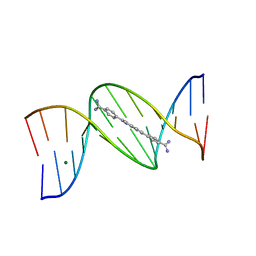 | | 5'-CGCGAATTCGCG-3' and an AT-specific binder (DB1884) complex | | Descriptor: | 4,4'-(1H-indole-2,6-diyl)dibenzenecarboximidamide, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-09-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8DSN
 
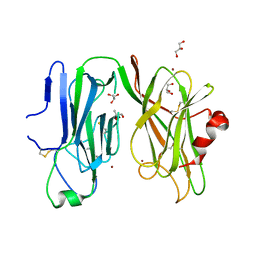 | | Peptidylglycine alpha hydroxylating monoxygenase, Q272A | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidylglycine alpha-amidating monooxygenase | | Authors: | Arias, R.J, Blackburn, N.J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New structures reveal flexible dynamics between the subdomains of peptidylglycine monooxygenase. Implications for an open to closed mechanism.
Protein Sci., 32, 2023
|
|
8DSJ
 
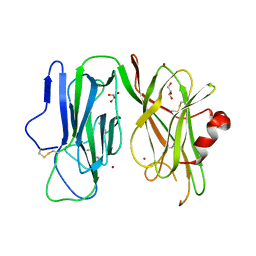 | | Peptidylglycine alpha hydroxylating monooxygenase anaerobic | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidylglycine alpha-amidating monooxygenase | | Authors: | Arias, R.J, Blackburn, N.J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New structures reveal flexible dynamics between the subdomains of peptidylglycine monooxygenase. Implications for an open to closed mechanism.
Protein Sci., 32, 2023
|
|
8E51
 
 | | Crystal Structure of Iridescent Shark Catfish Cadherin-23 EC1-2 and Protocadherin-15 EC1-2 | | Descriptor: | CALCIUM ION, Cadherin-23, POTASSIUM ION, ... | | Authors: | Scheib, E, Nisler, C.R, Sotomayor, M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Interpreting the Evolutionary Echoes of a Protein Complex Essential for Inner-Ear Mechanosensation.
Mol.Biol.Evol., 40, 2023
|
|
8DVN
 
 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
8C89
 
 | | SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab) | | Descriptor: | 17T2 Fab heavy chain, 17T2 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Modrego, A, Carlero, D, Bueno-Carrasco, M.T, Santiago, C, Carolis, C, Arranz, R, Blanco, J, Magri, G. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.41 Å) | | Cite: | A monoclonal antibody targeting a large surface of the receptor binding motif shows pan-neutralizing SARS-CoV-2 activity.
Nat Commun, 15, 2024
|
|
4A3X
 
 | | Structure of the N-terminal domain of the Epa1 adhesin (Epa1-Np) from the pathogenic yeast Candida glabrata, in complex with calcium and lactose | | Descriptor: | CALCIUM ION, EPA1P, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ielasi, F.S, Decanniere, K, Willaert, R.G. | | Deposit date: | 2011-10-05 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Epithelial Adhesin 1 (Epa1P) from the Human-Pathogenic Yeast Candida Glabrata : Structural and Functional Study of the Carbohydrate-Binding Domain
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8C3T
 
 | |
8C7M
 
 | |
6E1H
 
 | | Structure of 2:1 human Ptch1-Shh-N complex | | Descriptor: | CALCIUM ION, PALMITIC ACID, Protein patched homolog 1, ... | | Authors: | Qi, X, Li, X. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex.
Science, 362, 2018
|
|
8CVP
 
 | | Cereblon-DDB1 in the Apo form | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, ZINC ION | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8D7V
 
 | |
8D7X
 
 | |
8DDU
 
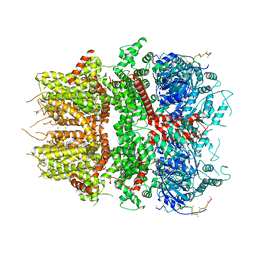 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state3 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDQ
 
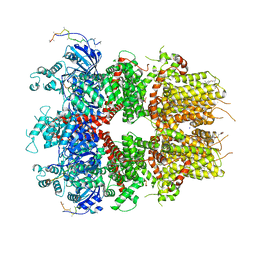 | | cryo-EM structure of TRPM3 ion channel in the presence of soluble Gbg, focused on channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8D80
 
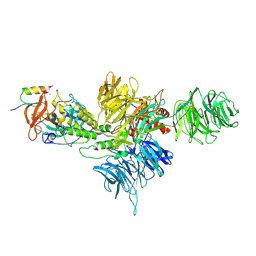 | | Cereblon~DDB1 bound to Iberdomide and Ikaros ZF1-2-3 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, DNA-binding protein Ikaros, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
