9BNR
 
 | | 4-(2-isothiocyanatoethyl)benzenesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | Macrophage migration inhibitory factor, N-[2-(4-sulfamoylphenyl)ethyl]methanethioamide, SULFATE ION | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
9BNQ
 
 | | N-(4-(isothiocyanatomethyl)phenyl)methanesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, N-{[4-(methanesulfonamido)phenyl]methyl}methanethioamide, ... | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
7OCL
 
 | |
3ZD1
 
 | | STRUCTURE OF THE TWO C-TERMINAL DOMAINS OF COMPLEMENT FACTOR H RELATED PROTEIN 2 | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H-RELATED PROTEIN 2 | | Authors: | Caesar, J.J.E, Goicoechea de Jorge, E, Pickering, M.C, Lea, S.M. | | Deposit date: | 2012-11-23 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dimerization of Complement Factor H-Related Proteins Modulates Complement Activation in Vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2W3I
 
 | | Crystal Structure of FXa in complex with 4,4-disubstituted pyrrolidine-1,2-dicarboxamide inhibitor 2 | | Descriptor: | (2R,4S)-N^1^-(4-chlorophenyl)-4-(2,4-difluorophenyl)-4-hydroxy-N^2^-(2-oxo-2H-1,3'-bipyridin-6'-yl)pyrrolidine-1,2-dicarboxamide, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Zhang, E, Mochalkin, I, Casimiro-Garcia, A, Van Huis, C.A. | | Deposit date: | 2008-11-12 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploration of 4,4-Disubstituted Pyrrolidine-1,2-Dicarboxamides as Potent, Orally Active Factor Xa Inhibitors with Extended Duration of Action.
Bioorg.Med.Chem., 17, 2009
|
|
6SVK
 
 | | human Myeloid-derived growth factor (MYDGF) | | Descriptor: | MALONATE ION, Myeloid-derived growth factor | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2019-09-18 | | Release date: | 2019-11-27 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Crystal structure and receptor-interacting residues of MYDGF - a protein mediating ischemic tissue repair.
Nat Commun, 10, 2019
|
|
8TN9
 
 | | Structural architecture of the acidic region of the B domain of coagulation factor V | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor V | | Authors: | Mohammed, B.M, Basore, K, Summers, B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2023-08-01 | | Release date: | 2023-10-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural architecture of the acidic region of the B domain of coagulation factor V.
J.Thromb.Haemost., 22, 2024
|
|
6SVL
 
 | |
7KXY
 
 | |
3MKP
 
 | | Crystal structure of 1K1 mutant of Hepatocyte Growth Factor/Scatter Factor fragment NK1 in complex with heparin | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Gherardi, E, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Engineering a fragment of Hepatocyte Growth Factor/Scatter Factor for tissue and organ regeneration
To be Published
|
|
1K6Q
 
 | | Crystal structure of antibody Fab fragment D3 | | Descriptor: | immunoglobulin Fab D3, heavy chain, light chain | | Authors: | Faelber, K, Kelley, R.F, Kirchhofer, D, Muller, Y.A. | | Deposit date: | 2001-10-17 | | Release date: | 2002-04-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of murine Fab D3 at 2.4 A resolution in comparison with the humanised Fab D3h44 (1.85A) provides structural insight into the humanisation process of the D3 anti-tissue factor antibody
To be Published
|
|
7OCM
 
 | | K1K1H6, a potent recombinant minimal hepatocyte growth factor/scatter factor mimic | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hepatocyte growth factor alpha chain,Hepatocyte growth factor alpha chain | | Authors: | de Jonge, H, de Nola, G, Gherardi, E. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimerization of kringle 1 domain from hepatocyte growth factor/scatter factor provides a potent MET receptor agonist.
Life Sci Alliance, 5, 2022
|
|
5MIV
 
 | | G307E variant of murine Apoptosis Inducing Factor in complex with NAD+ | | Descriptor: | Apoptosis-inducing factor 1, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sorrentino, L, Cossu, F, Aliverti, A, Milani, M, Mastrangelo, E. | | Deposit date: | 2016-11-29 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural bases of the altered catalytic properties of a pathogenic variant of apoptosis inducing factor.
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5MIU
 
 | | G307E variant of Murine Apoptosis Inducing Factor (oxidized state) | | Descriptor: | Apoptosis-inducing factor 1, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Sorrentino, L, Cossu, F, Milani, M, Aliverti, A, Mastrangelo, E. | | Deposit date: | 2016-11-29 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural bases of the altered catalytic properties of a pathogenic variant of apoptosis inducing factor.
Biochem. Biophys. Res. Commun., 490, 2017
|
|
8DEA
 
 | |
4DTG
 
 | | Hemostatic effect of a monoclonal antibody mAb 2021 blocking the interaction between FXa and TFPI in a rabbit hemophilia model | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Humanized recombinant FAB fragment, ... | | Authors: | Svensson, L.A, Breinholt, J, Krogh, B.O, Hilden, I. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hemostatic effect of a monoclonal antibody mAb 2021 blocking the interaction between FXa and TFPI in a rabbit hemophilia model.
Blood, 119, 2012
|
|
7V1N
 
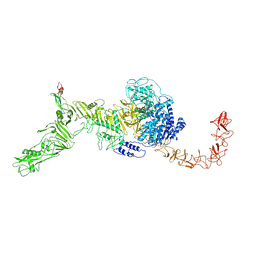 | | Structure of the Clade 2 C. difficile TcdB in complex with its receptor TFPI | | Descriptor: | Isoform Beta of Tissue factor pathway inhibitor, Toxin B | | Authors: | Luo, J, Yang, Q, Zhang, X, Zhang, Y, Wan, L, Li, Y, Tao, L. | | Deposit date: | 2021-08-05 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile.
Cell, 185, 2022
|
|
8DG6
 
 | |
8D95
 
 | |
1B8M
 
 | | BRAIN DERIVED NEUROTROPHIC FACTOR, NEUROTROPHIN-4 | | Descriptor: | PROTEIN (BRAIN DERIVED NEUROTROPHIC FACTOR), PROTEIN (NEUROTROPHIN-4) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
5CXT
 
 | |
1BOE
 
 | | STRUCTURE OF THE IGF BINDING DOMAIN OF THE INSULIN-LIKE GROWTH FACTOR-BINDING PROTEIN-5 (IGFBP-5): IMPLICATIONS FOR IGF AND IGF-I RECEPTOR INTERACTIONS | | Descriptor: | PROTEIN (INSULIN-LIKE GROWTH FACTOR-BINDING PROTEIN-5 (IGFBP-5)) | | Authors: | Kalus, W, Zweckstetter, M, Renner, C, Sanchez, Y, Georgescu, J, Grol, M, Demuth, D, Schumacherdony, C, Lang, K, Holak, T.H. | | Deposit date: | 1998-07-30 | | Release date: | 1998-12-16 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the IGF-binding domain of the insulin-like growth factor-binding protein-5 (IGFBP-5): implications for IGF and IGF-I receptor interactions.
EMBO J., 17, 1998
|
|
3ZD2
 
 | | THE STRUCTURE OF THE TWO N-TERMINAL DOMAINS OF COMPLEMENT FACTOR H RELATED PROTEIN 1 SHOWS FORMATION OF A NOVEL DIMERISATION INTERFACE | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H-RELATED PROTEIN 1 | | Authors: | Caesar, J.J.E, Goicoechea de Jorge, E, Pickering, M.C, Lea, S.M. | | Deposit date: | 2012-11-23 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Dimerization of Complement Factor H-Related Proteins Modulates Complement Activation in Vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2LPN
 
 | | Solution Structure of N-Terminal domain of human Conserved Dopamine Neurotrophic Factor (CDNF) | | Descriptor: | Cerebral dopamine neurotrophic factor | | Authors: | Latge, C, Cabral, K.M.S, Foguel, D, Pires, J.R.M, Almeida, M.S. | | Deposit date: | 2012-02-15 | | Release date: | 2013-02-20 | | Last modified: | 2013-05-22 | | Method: | SOLUTION NMR | | Cite: | (1)H-, (13)C- and (15)N-NMR assignment of the N-terminal domain of human cerebral dopamine neurotrophic factor (CDNF).
Biomol.Nmr Assign., 7, 2013
|
|
3HMT
 
 | |
