6ICV
 
 | | Structure of SETD3 bound to SAH and unmodified actin | | Descriptor: | Actin, cytoplasmic 1, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Guo, Q, Liao, S, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into SETD3-mediated histidine methylation on beta-actin.
Elife, 8, 2019
|
|
6MBJ
 
 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, P21 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6MBK
 
 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, First P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, Actin peptide, GLYCEROL, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6MBL
 
 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, Second P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6MBO
 
 | | GLP Methyltransferase with Inhibitor EML741-P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-7-methoxy-N-[1-(propan-2-yl)piperidin-4-yl]-8-[3-(pyrrolidin-1-yl)propoxy]-3H-1,4-benzodiazepin-5-amine, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Discovery of a Novel Chemotype of Histone Lysine Methyltransferase EHMT1/2 (GLP/G9a) Inhibitors: Rational Design, Synthesis, Biological Evaluation, and Co-crystal Structure.
J. Med. Chem., 62, 2019
|
|
6MBP
 
 | | GLP Methyltransferase with Inhibitor EML741- P3121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-7-methoxy-N-[1-(propan-2-yl)piperidin-4-yl]-8-[3-(pyrrolidin-1-yl)propoxy]-3H-1,4-benzodiazepin-5-amine, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Discovery of a Novel Chemotype of Histone Lysine Methyltransferase EHMT1/2 (GLP/G9a) Inhibitors: Rational Design, Synthesis, Biological Evaluation, and Co-crystal Structure.
J. Med. Chem., 62, 2019
|
|
6AGO
 
 | | Crystal structure of MRG15-ASH1L Histone methyltransferase complex | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, Mortality factor 4 like 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Lee, Y, Song, J. | | Deposit date: | 2018-08-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis of MRG15-Mediated Activation of the ASH1L Histone Methyltransferase by Releasing an Autoinhibitory Loop.
Structure, 27, 2019
|
|
6A5M
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 2 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5N
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5K
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 1 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CHG
 
 | | Crystal structure of the yeast COMPASS catalytic module | | Descriptor: | H3, Histone-lysine N-methyltransferase, H3 lysine-4 specific, ... | | Authors: | Hsu, P.L, Li, H, Zheng, N. | | Deposit date: | 2018-02-22 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal Structure of the COMPASS H3K4 Methyltransferase Catalytic Module.
Cell, 174, 2018
|
|
6CEN
 
 | | Crystal Structure of WHSC1L1 in Complex with Inhibitor PEP21 | | Descriptor: | ACE-GLY-VAL-NLE-ARG-ILE-NH2, Histone-lysine N-methyltransferase NSD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Boriack-Sjodin, P.A, Swinger, K, Farrow, N.A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Identification of a peptide inhibitor for the histone methyltransferase WHSC1.
PLoS ONE, 13, 2018
|
|
6CBY
 
 | | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF9975 | | Descriptor: | N-lysine methyltransferase SMYD2, ZINC ION, [3-(4-amino-6-methyl-1H-imidazo[4,5-c]pyridin-1-yl)-3-methylazetidin-1-yl][1-({1-[(1R)-cyclohept-2-en-1-yl]piperidin-4-yl}methyl)-1H-pyrrol-3-yl]methanone | | Authors: | ZENG, H, DONG, A, Hutchinson, A, Seitova, A, TATLOCK, J, KUMPF, R, OWEN, A, TAYLOR, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-05 | | Release date: | 2018-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Selective, Small-Molecule Co-Factor Binding Site Inhibition of a Su(var)3-9, Enhancer of Zeste, Trithorax Domain Containing Lysine Methyltransferase.
J.Med.Chem., 62, 2019
|
|
6CBX
 
 | | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF1497 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-lysine methyltransferase SMYD2, ... | | Authors: | ZENG, H, DONG, A, Hutchinson, A, Seitova, A, TATLOCK, J, KUMPF, R, OWEN, A, TAYLOR, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF1497
to be published
|
|
6FND
 
 | | Crystal structure of Toxoplasma gondii AKMT | | Descriptor: | 1,2-ETHANEDIOL, Apical complex lysine methyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pivovarova, Y, Dong, G. | | Deposit date: | 2018-02-02 | | Release date: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of a Novel Dimeric SET Domain Methyltransferase that Regulates Cell Motility.
J. Mol. Biol., 430, 2018
|
|
6C24
 
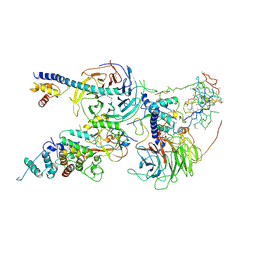 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-06 | | Release date: | 2018-01-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
6C23
 
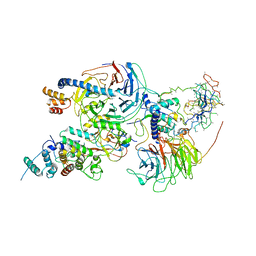 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-05 | | Release date: | 2018-01-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
6BX3
 
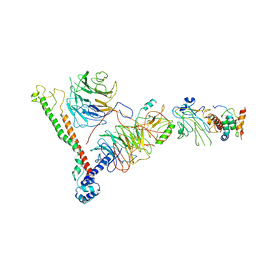 | | Structure of histone H3k4 methyltransferase | | Descriptor: | COMPASS component BRE2, COMPASS component SDC1, COMPASS component SPP1, ... | | Authors: | Skiniotis, G, Qu, Q.H. | | Deposit date: | 2017-12-16 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex.
Cell, 174, 2018
|
|
6BOZ
 
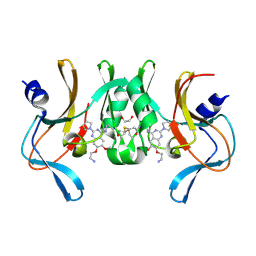 | | Structure of human SETD8 in complex with covalent inhibitor MS4138 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)prop-2-enamide, N-lysine methyltransferase KMT5A | | Authors: | Babault, N, Anqi, M, Jin, J. | | Deposit date: | 2017-11-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
6BP4
 
 | | Structure of the S. pombe Clr4 catalytic domain bound to SAM | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Currie, M.A, Moazed, D. | | Deposit date: | 2017-11-21 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7701 Å) | | Cite: | Automethylation-induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability.
Nature, 560, 2018
|
|
6BOX
 
 | | Structure of the S. pombe Clr4 catalytic domain bound to SAH | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Currie, M.A, Moazed, D. | | Deposit date: | 2017-11-21 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Automethylation-induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability.
Nature, 560, 2018
|
|
5YLT
 
 | | Crystal structure of SET7/9 in complex with a cyproheptadine derivative | | Descriptor: | 2-(1-methylpiperidin-4-ylidene)tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3(8),4,6,9,12,14-heptaen-6-ol, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Hirano, T, Fujiwara, T, Niwa, H, Hirano, M, Ohira, K, Okazaki, Y, Sato, S, Umehara, T, Maemoto, Y, Ito, A, Yoshida, M, Kagechika, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of Novel Inhibitors for Histone Methyltransferase SET7/9 based on Cyproheptadine.
ChemMedChem, 13, 2018
|
|
5YJO
 
 | |
4W2R
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
6B3W
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit,Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
