4RZ7
 
 | | Crystal Structure of PVX_084705 with bound PCI32765 | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, UNKNOWN ATOM OR ION, cGMP-dependent protein kinase, ... | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal Structure of PVX_084705 with bound PCI32765
To be Published
|
|
4RYV
 
 | | Crystal structure of yellow lupin LLPR-10.1A protein in complex with trans-zeatin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Protein LLPR-10.1A, SULFATE ION | | Authors: | Dolot, R, Michalska, K, Sliwiak, J, Bujacz, G, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystallographic and CD probing of ligand-induced conformational changes in a plant PR-10 protein.
J.Struct.Biol., 193, 2016
|
|
5KSI
 
 | | Crystal structure of deoxygenated hemoglobin in complex with sphingosine phosphate and 2,3-Bisphosphoglycerate | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Hemoglobin subunit alpha, ... | | Authors: | Ahmed, M.H, Safo, M.K, Xia, Y. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Insight of Sphingosine 1-Phosphate-Mediated Pathogenic Metabolic Reprogramming in Sickle Cell Disease.
Sci Rep, 7, 2017
|
|
5KWY
 
 | | Structure of human NPC1 middle lumenal domain bound to NPC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLEST-5-EN-3-YL HYDROGEN SULFATE, Epididymal secretory protein E1, ... | | Authors: | Li, X. | | Deposit date: | 2016-07-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7MXY
 
 | | Cryo-EM structure of PfFNT-inhibitor complex | | Descriptor: | (Z)-4,4,5,5,5-pentakis(fluoranyl)-1-(4-methoxy-2-oxidanyl-phenyl)-3-oxidanyl-pent-2-en-1-one, Formate-nitrite transporter | | Authors: | Yu, E.W, Su, C, Lyu, M. | | Deposit date: | 2021-05-19 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structural basis of transport and inhibition of the Plasmodium falciparum transporter PfFNT
EMBO Rep, 22, 2021
|
|
7N9D
 
 | |
4RMD
 
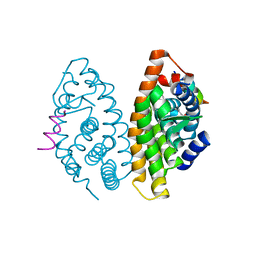 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with 9cUAB110 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-8-[3-cyclopropyl-2-(3-methylbutyl)cyclohex-2-en-1-ylidene]-3,7-dimethylocta-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Muccio, D.D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
4RMC
 
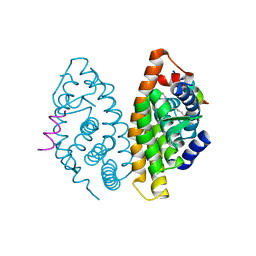 | | Crystal Structure of human retinoid X receptor alpha-ligand binding domain complex with 9cUAB76 and the coactivator peptide GRIP-1 | | Descriptor: | (3S,7S,8E)-8-[3-ethyl-2-(3-methylbutyl)cyclohex-2-en-1-ylidene]-3,7-dimethyloctanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Muccio, D.D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
7MFZ
 
 | |
7MFX
 
 | |
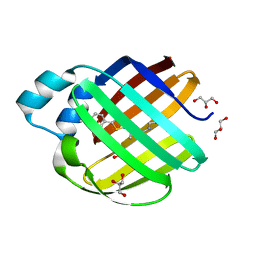
7MFY
 
 | |
4TTV
 
 | | Crystal structure of human ThrRS complexing with a bioengineered macrolide BC194 | | Descriptor: | (1R,2R)-2-[(2S,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadec-6-en-2-yl]cyclobutanecarboxylic acid, Threonine--tRNA ligase, cytoplasmic, ... | | Authors: | Fang, P, Guo, M. | | Deposit date: | 2014-06-23 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Aminoacyl-tRNA synthetase dependent angiogenesis revealed by a bioengineered macrolide inhibitor.
Sci Rep, 5, 2015
|
|
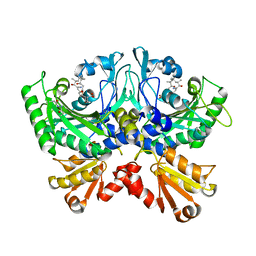
5J9Y
 
 | | EGFR-T790M in complex with pyrazolopyrimidine inhibitor 1b | | Descriptor: | (R)-1-(3-(4-amino-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl)prop-2-en-1-one, Epidermal growth factor receptor | | Authors: | Becker, C, Engel, J, Rauh, D. | | Deposit date: | 2016-04-11 | | Release date: | 2016-08-17 | | Last modified: | 2016-09-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insight into the Inhibition of Drug-Resistant Mutants of the Receptor Tyrosine Kinase EGFR.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
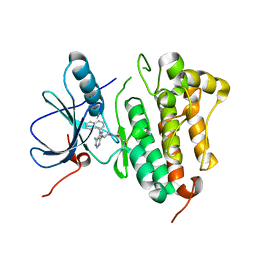
5J14
 
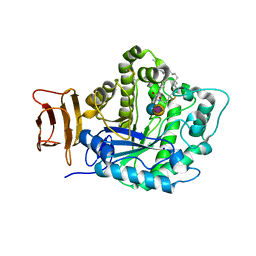 | | Crystal structure of endoglycoceramidase I from Rhodococ-cus equi in complex with GM3 | | Descriptor: | N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Putative secreted endoglycosylceramidase, ... | | Authors: | Chen, L. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.915 Å) | | Cite: | Structural Insights into the Broad Substrate Specificity of a Novel Endoglycoceramidase I Belonging to a New Subfamily of GH5 Glycosidases
J. Biol. Chem., 292, 2017
|
|
4TW7
 
 | | The Fk1 domain of FKBP51 in complex with iFit4 | | Descriptor: | (1R)-3-(3,4-dimethoxyphenyl)-1-{3-[2-(morpholin-4-yl)ethoxy]phenyl}propyl (2S)-1-[(2S)-2-[(1S)-cyclohex-2-en-1-yl]-2-(3,4,5-trimethoxyphenyl)acetyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
7N42
 
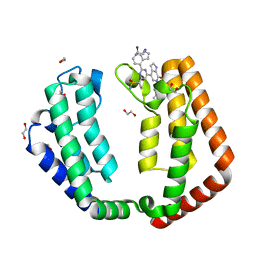 | | Crystal structure of the tandem bromodomain of human TAF1 (TAF1-T) bound to ZS1-681 | | Descriptor: | 1,2-ETHANEDIOL, 6-(but-3-en-1-yl)-4-[6-{1-[(R)-S-methanesulfonimidoyl]cyclopropyl}-2-(1H-pyrrolo[2,3-b]pyridin-4-yl)pyrimidin-4-yl]-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, DIMETHYL SULFOXIDE, ... | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the tandem bromodomain of human TAF1 (TAF1-T) bound to ZS1-681
To Be Published
|
|
4TUN
 
 | |
5LNU
 
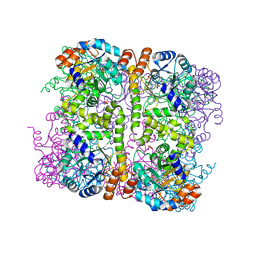 | | Crystal structure of Arabidopsis thaliana Pdx1-I320 complex | | Descriptor: | (4~{S})-4-azanyl-5-oxidanyl-pent-1-en-3-one, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2017-02-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LCJ
 
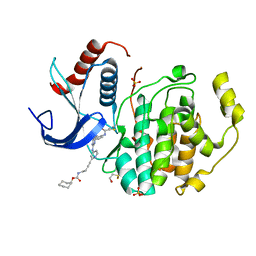 | | In-Gel Activity-Based Protein Profiling of a Clickable Covalent Erk 1/2 Inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, [(1~{R},4~{Z})-cyclooct-4-en-1-yl] ~{N}-[4-[4-[[5-chloranyl-4-[[2-(propanoylamino)phenyl]amino]pyrimidin-2-yl]amino]pyridin-2-yl]but-3-ynyl]carbamate | | Authors: | O'Reilly, M, Wright, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-20 | | Last modified: | 2016-08-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | In-gel activity-based protein profiling of a clickable covalent ERK1/2 inhibitor.
Mol Biosyst, 12, 2016
|
|
7MK5
 
 | | Crystal structure of Escherichia coli ClpP covalently inhibited by clipibicyclene | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-[(1E)-3-{[(2E,4E,6E,8S)-8-hydroxy-4-methyldeca-2,4,6-trienoyl]amino}-3-oxoprop-1-en-1-yl]azete-1(2H)-carboxylic acid, ACETATE ION, ... | | Authors: | Culp, E.J, Sychantha, D, Hobson, C, Pawlowski, A.J, Prehna, G, Wright, G.D. | | Deposit date: | 2021-04-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | ClpP inhibitors are produced by a widespread family of bacterial gene clusters.
Nat Microbiol, 7, 2022
|
|
4TW8
 
 | | The Fk1-Fk2 domains of FKBP52 in complex with iFit-FL | | Descriptor: | 2-(5-{[({3-[(1R)-1-[({(2S)-1-[(2S)-2-[(1S)-cyclohex-2-en-1-yl]-2-(3,4,5-trimethoxyphenyl)acetyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetyl)amino]methyl}-6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic acid, Peptidyl-prolyl cis-trans isomerase FKBP4 | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
5JZE
 
 | | Erve virus viral OTU domain protease in complex with mouse ISG15 | | Descriptor: | CITRATE ANION, RNA-dependent RNA polymerase, Ubiquitin-like protein ISG15, ... | | Authors: | Deaton, M.K, Dzimianski, J.V, Pegan, S.D. | | Deposit date: | 2016-05-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Biochemical and Structural Insights into the Preference of Nairoviral DeISGylases for Interferon-Stimulated Gene Product 15 Originating from Certain Species.
J.Virol., 90, 2016
|
|
5LNW
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-I320-G3P complex | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, GLYCEROL, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
7MTU
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P221 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-13 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P221
To Be Published
|
|
4UMM
 
 | | The Cryo-EM structure of the palindromic DNA-bound USP-EcR nuclear receptor reveals an asymmetric organization with allosteric domain positioning | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, 5'-D(*CP*AP*AP*GP*GP*GP*TP*TP*CP*AP*AP*TP*GP*CP *AP*CP*TP*TP*GP*TP)-3', 5'-D(*DGP*AP*CP*AP*AP*GP*TP*GP*CP*AP*TP*TP*GP*DAP *AP*CP*CP*CP*TP*T)-3', ... | | Authors: | Maletta, M, Orlov, I, Moras, D, Billas, I.M.L, Klaholz, B.P. | | Deposit date: | 2014-05-19 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | The Palindromic DNA-Bound Usp-Ecr Nuclear Receptor Adopts an Asymmetric Organization with Allosteric Domain Positioning.
Nat.Commun., 5, 2014
|
|
